Pathway analysis from lists of microRNAs: common pitfalls and alternative strategy
- PMID: 25800743
- PMCID: PMC4402548
- DOI: 10.1093/nar/gkv249
Pathway analysis from lists of microRNAs: common pitfalls and alternative strategy
Abstract
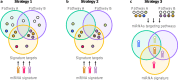
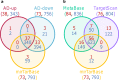
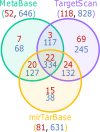
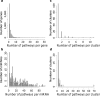
MicroRNAs (miRNAs) are involved in the regulation of gene expression at a post-transcriptional level. As such, monitoring miRNA expression has been increasingly used to assess their role in regulatory mechanisms of biological processes. In large scale studies, once miRNAs of interest have been identified, the target genes they regulate are often inferred using algorithms or databases. A pathway analysis is then often performed in order to generate hypotheses about the relevant biological functions controlled by the miRNA signature. Here we show that the method widely used in scientific literature to identify these pathways is biased and leads to inaccurate results. In addition to describing the bias and its origin we present an alternative strategy to identify potential biological functions specifically impacted by a miRNA signature. More generally, our study exemplifies the crucial need of relevant negative controls when developing, and using, bioinformatics methods.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Pasquinelli A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

