Diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis
- PMID: 25800755
- PMCID: PMC4397280
- DOI: 10.1200/JCO.2014.57.7080
Diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis
Abstract
Purpose: Current diagnostic tests for diffuse large B-cell lymphoma use the updated WHO criteria based on biologic, morphologic, and clinical heterogeneity. We propose a refined classification system based on subset-specific B-cell-associated gene signatures (BAGS) in the normal B-cell hierarchy, hypothesizing that it can provide new biologic insight and diagnostic and prognostic value.
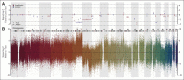
Patients and methods: We combined fluorescence-activated cell sorting, gene expression profiling, and statistical modeling to generate BAGS for naive, centrocyte, centroblast, memory, and plasmablast B cells from normal human tonsils. The impact of BAGS-assigned subtyping was analyzed using five clinical cohorts (treated with cyclophosphamide, doxorubicin, vincristine, and prednisone [CHOP], n = 270; treated with rituximab plus CHOP [R-CHOP], n = 869) gathered across geographic regions, time eras, and sampling methods. The analysis estimated subtype frequencies and drug-specific resistance and included a prognostic meta-analysis of patients treated with first-line R-CHOP therapy.
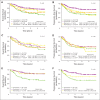
Results: Similar BAGS subtype frequencies were assigned across 1,139 samples from five different cohorts. Among R-CHOP-treated patients, BAGS assignment was significantly associated with overall survival and progression-free survival within the germinal center B-cell-like subclass; the centrocyte subtype had a superior prognosis compared with the centroblast subtype. In agreement with the observed therapeutic outcome, centrocyte subtypes were estimated as being less resistant than the centroblast subtype to doxorubicin and vincristine. The centroblast subtype had a complex genotype, whereas the centrocyte subtype had high TP53 mutation and insertion/deletion frequencies and expressed LMO2, CD58, and stromal-1-signature and major histocompatibility complex class II-signature genes, which are known to have a positive impact on prognosis.
Conclusion: Further development of a diagnostic platform using BAGS-assigned subtypes may allow pathogenetic studies to improve disease management.
© 2015 by American Society of Clinical Oncology.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest are found in the article online at
Figures





References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Andréasson U, Dictor M, Jerkeman M, et al. Identification of molecular targets associated with transformed diffuse large B cell lymphoma using highly purified tumor cells. Am J Hematol. 2009;84:803–808. - PubMed
-
- Kjeldsen MK, Perez-Andres M, Schmitz A, et al. Multiparametric flow cytometry for identification and fluorescence activated cell sorting of five distinct B-cell subpopulations in normal tonsil tissue. Am J Clin Pathol. 2011;136:960–969. - PubMed
-
- Perez-Andres M, Paiva B, Nieto WG, et al. Human peripheral blood B-cell compartments: A crossroad in B-cell traffic. Cytometry B Clin Cytom. 2010;78(suppl 1):S47–S60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

