UHRF1 is a sensor for DNA interstrand crosslinks and recruits FANCD2 to initiate the Fanconi anemia pathway
- PMID: 25801034
- PMCID: PMC4386029
- DOI: 10.1016/j.celrep.2015.02.053
UHRF1 is a sensor for DNA interstrand crosslinks and recruits FANCD2 to initiate the Fanconi anemia pathway
Abstract
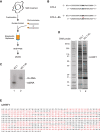
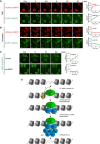
The Fanconi anemia (FA) pathway is critical for the cellular response to toxic DNA interstrand crosslinks (ICLs). Using a biochemical purification strategy, we identified UHRF1 as a protein that specifically interacts with ICLs in vitro and in vivo. Reduction of cellular levels of UHRF1 by RNAi attenuates the FA pathway and sensitizes cells to mitomycin C. Knockdown cells display a drastic reduction in FANCD2 foci formation. Using live-cell imaging, we observe that UHRF1 is rapidly recruited to chromatin in response to DNA crosslinking agents and that this recruitment both precedes and is required for the recruitment of FANCD2 to ICLs. Based on these results, we describe a mechanism of ICL sensing and propose that UHRF1 is a critical factor that binds to ICLs. In turn, this binding is necessary for the subsequent recruitment of FANCD2, which allows the DNA repair process to initiate.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. - PubMed
-
- Bostick M., Kim J.K., Estève P.O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. - PubMed
-
- Ciccia A., Ling C., Coulthard R., Yan Z., Xue Y., Meetei A.R., Laghmani H., Joenje H., McDonald N., de Winter J.P. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

