Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9
- PMID: 25803037
- PMCID: PMC4372442
- DOI: 10.1371/journal.pone.0120396
Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9
Abstract
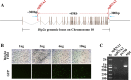
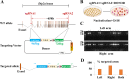
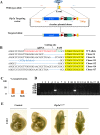
ZFN, TALENs and CRISPR/Cas9 system have been used to generate point mutations and large fragment deletions and insertions in genomic modifications. CRISPR/Cas9 system is the most flexible and fast developing technology that has been extensively used to make mutations in all kinds of organisms. However, the most mutations reported up to date are small insertions and deletions. In this report, CRISPR/Cas9 system was used to make large DNA fragment deletions and insertions, including entire Dip2a gene deletion, about 65kb in size, and β-galactosidase (lacZ) reporter gene insertion of larger than 5kb in mouse. About 11.8% (11/93) are positive for 65kb deletion from transfected and diluted ES clones. High targeting efficiencies in ES cells were also achieved with G418 selection, 46.2% (12/26) and 73.1% (19/26) for left and right arms respectively. Targeted large fragment deletion efficiency is about 21.4% of live pups or 6.0% of injected embryos. Targeted insertion of lacZ reporter with NEO cassette showed 27.1% (13/48) of targeting rate by ES cell transfection and 11.1% (2/18) by direct zygote injection. The procedures have bypassed in vitro transcription by directly co-injection of zygotes or co-transfection of embryonic stem cells with circular plasmid DNA. The methods are technically easy, time saving, and cost effective in generating mouse models and will certainly facilitate gene function studies.
Conflict of interest statement
Figures

References
-
- Mizuno S, Dinh TT, Kato K, Mizuno-Iijima S, Tanimoto Y, Daitoku Y, et al. Simple generation of albino C57BL/6J mice with G291T mutation in the tyrosinase gene by the CRISPR/Cas9 system. Mammalian genome: official journal of the International Mammalian Genome Society. 2014;25(7–8):327–334. - PubMed
-
- Capecchi MR. Generating mice with targeted mutations. Nature medicine. 2001;7(10):1086–1090. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

