Prevention of biofilm formation and removal of existing biofilms by extracellular DNases of Campylobacter jejuni
- PMID: 25803828
- PMCID: PMC4372405
- DOI: 10.1371/journal.pone.0121680
Prevention of biofilm formation and removal of existing biofilms by extracellular DNases of Campylobacter jejuni
Abstract
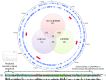
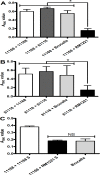
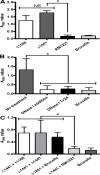
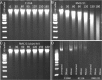
The fastidious nature of the foodborne bacterial pathogen Campylobacter jejuni contrasts with its ability to survive in the food chain. The formation of biofilms, or the integration into existing biofilms by C. jejuni, is thought to contribute to food chain survival. As extracellular DNA (eDNA) has previously been proposed to play a role in C. jejuni biofilms, we have investigated the role of extracellular DNases (eDNases) produced by C. jejuni in biofilm formation. A search of 2791 C. jejuni genomes highlighted that almost half of C. jejuni genomes contains at least one eDNase gene, but only a minority of isolates contains two or three of these eDNase genes, such as C. jejuni strain RM1221 which contains the cje0256, cje0566 and cje1441 eDNase genes. Strain RM1221 did not form biofilms, whereas the eDNase-negative strains NCTC 11168 and 81116 did. Incubation of pre-formed biofilms of NCTC 11168 with live C. jejuni RM1221 or with spent medium from a RM1221 culture resulted in removal of the biofilm. Inactivation of the cje1441 eDNase gene in strain RM1221 restored biofilm formation, and made the mutant unable to degrade biofilms of strain NCTC 11168. Finally, C. jejuni strain RM1221 was able to degrade genomic DNA from C. jejuni NCTC 11168, 81116 and RM1221, whereas strain NCTC 11168 and the RM1221 cje1441 mutant were unable to do so. This was mirrored by an absence of eDNA in overnight cultures of C. jejuni RM1221. This suggests that the activity of eDNases in C. jejuni affects biofilm formation and is not conducive to a biofilm lifestyle. These eDNases do however have a potential role in controlling biofilm formation by C. jejuni strains in food chain relevant environments.
Conflict of interest statement
Figures

References
-
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science 2002;295: 1487 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

