Spontaneous dominant mutations in chlamydomonas highlight ongoing evolution by gene diversification
- PMID: 25804537
- PMCID: PMC4558696
- DOI: 10.1105/tpc.15.00010
Spontaneous dominant mutations in chlamydomonas highlight ongoing evolution by gene diversification
Abstract
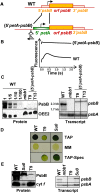
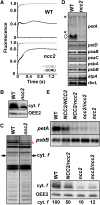
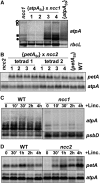
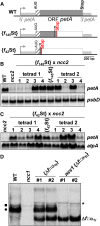
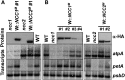
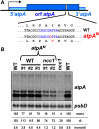
We characterized two spontaneous and dominant nuclear mutations in the unicellular alga Chlamydomonas reinhardtii, ncc1 and ncc2 (for nuclear control of chloroplast gene expression), which affect two octotricopeptide repeat (OPR) proteins encoded in a cluster of paralogous genes on chromosome 15. Both mutations cause a single amino acid substitution in one OPR repeat. As a result, the mutated NCC1 and NCC2 proteins now recognize new targets that we identified in the coding sequences of the chloroplast atpA and petA genes, respectively. Interaction of the mutated proteins with these targets leads to transcript degradation; however, in contrast to the ncc1 mutation, the ncc2 mutation requires on-going translation to promote the decay of the petA mRNA. Thus, these mutants reveal a mechanism by which nuclear factors act on chloroplast mRNAs in Chlamydomonas. They illustrate how diversifying selection can allow cells to adapt the nuclear control of organelle gene expression to environmental changes. We discuss these data in the wider context of the evolution of regulation by helical repeat proteins.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures



Comment in
-
Twice as NCC: Two Octotricopeptide Repeat Proteins and the Regulation of Chloroplast Gene Expression.Plant Cell. 2015 Apr;27(4):947. doi: 10.1105/tpc.15.00211. Epub 2015 Mar 24. Plant Cell. 2015. PMID: 25804538 Free PMC article. No abstract available.
Similar articles
-
MDA1, a nucleus-encoded factor involved in the stabilization and processing of the atpA transcript in the chloroplast of Chlamydomonas.Plant J. 2019 Jun;98(6):1033-1047. doi: 10.1111/tpj.14300. Epub 2019 Apr 4. Plant J. 2019. PMID: 30809889
-
Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts.Plant J. 2011 Sep;67(6):1055-66. doi: 10.1111/j.1365-313X.2011.04657.x. Epub 2011 Jul 18. Plant J. 2011. PMID: 21623973
-
Chloroplast biogenesis involves spatial coordination of nuclear and organellar gene expression in Chlamydomonas.Plant Physiol. 2024 Sep 2;196(1):112-123. doi: 10.1093/plphys/kiae256. Plant Physiol. 2024. PMID: 38709497 Free PMC article.
-
The unicellular green alga Chlamydomonas reinhardtii as an experimental system to study chloroplast RNA metabolism.Naturwissenschaften. 2000 Mar;87(3):97-107. doi: 10.1007/s001140050686. Naturwissenschaften. 2000. PMID: 10798194 Review.
-
Loss of chloroplast ClpP elicits an autophagy-like response in Chlamydomonas.Autophagy. 2014 Sep;10(9):1685-6. doi: 10.4161/auto.29960. Epub 2014 Jul 17. Autophagy. 2014. PMID: 25046108 Free PMC article. Review.
Cited by
-
The OPR Protein MTHI1 Controls the Expression of Two Different Subunits of ATP Synthase CFo in Chlamydomonas reinhardtii.Plant Cell. 2020 Apr;32(4):1179-1203. doi: 10.1105/tpc.19.00770. Epub 2020 Jan 27. Plant Cell. 2020. PMID: 31988263 Free PMC article.
-
FASTKD1 and FASTKD4 have opposite effects on expression of specific mitochondrial RNAs, depending upon their endonuclease-like RAP domain.Nucleic Acids Res. 2017 Jun 2;45(10):6135-6146. doi: 10.1093/nar/gkx164. Nucleic Acids Res. 2017. PMID: 28335001 Free PMC article.
-
CCS2, an Octatricopeptide-Repeat Protein, Is Required for Plastid Cytochrome c Assembly in the Green Alga Chlamydomonas reinhardtii.Front Plant Sci. 2017 Aug 3;8:1306. doi: 10.3389/fpls.2017.01306. eCollection 2017. Front Plant Sci. 2017. PMID: 28824661 Free PMC article.
-
Polycytidylation of mitochondrial mRNAs in Chlamydomonas reinhardtii.Nucleic Acids Res. 2017 Dec 15;45(22):12963-12973. doi: 10.1093/nar/gkx903. Nucleic Acids Res. 2017. PMID: 29244187 Free PMC article.
-
Comparative genomics of Chlamydomonas.Plant Cell. 2021 May 31;33(4):1016-1041. doi: 10.1093/plcell/koab026. Plant Cell. 2021. PMID: 33793842 Free PMC article.
References
-
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552. - PubMed
-
- Anisimova M., Bielawski J.P., Yang Z. (2002). Accuracy and power of bayes prediction of amino acid sites under positive selection. Mol. Biol. Evol. 19: 950–958. - PubMed
-
- Barkan A., Goldschmidt-Clermont M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

