A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation
- PMID: 25805135
- PMCID: PMC4456036
- DOI: 10.1016/j.devcel.2015.01.028
A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation
Abstract
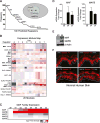
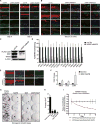
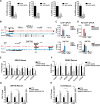
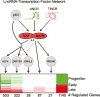
Progenitor differentiation requires remodeling of genomic expression; however, in many tissues, such as epidermis, the spectrum of remodeled genes and the transcription factors (TFs) that control them are not fully defined. We performed kinetic transcriptome analysis during regeneration of differentiated epidermis and identified gene sets enriched in progenitors (594 genes), in early (159 genes), and in late differentiation (387 genes). Module mapping of 1,046 TFs identified MAF and MAFB as necessary and sufficient for progenitor differentiation. MAF:MAFB regulated 393 genes altered in this setting. Integrative analysis identified ANCR and TINCR lncRNAs as essential upstream MAF:MAFB regulators. ChIP-seq analysis demonstrated MAF:MAFB binding to known epidermal differentiation TF genes whose expression they controlled, including GRHL3, ZNF750, KLF4, and PRDM1. Each of these TFs rescued expression of specific MAF:MAFB target gene subsets in the setting of MAF:MAFB loss, indicating they act downstream of MAF:MAFB. A lncRNA-TF network is thus essential for epidermal differentiation.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Dynamic networking for epidermal differentiation.Dev Cell. 2015 Mar 23;32(6):661-2. doi: 10.1016/j.devcel.2015.03.006. Dev Cell. 2015. PMID: 25805131
References
-
- Aziz A, Soucie E, Sarrazin S, Sieweke MH. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326:867–871. - PubMed
-
- Birnbaum RY, Zvulunov A, Hallel-Halevy D, Cagnano E, Finer G, Ofir R, Geiger D, Silberstein E, Feferman Y, Birk OS. Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat Genet. 2006;38:749–751. - PubMed
-
- Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

