A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity
- PMID: 25805733
- PMCID: PMC4453579
- DOI: 10.1128/mBio.02368-14
A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity
Abstract
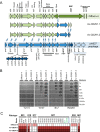
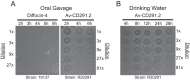
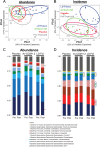
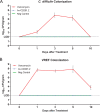
Clostridium difficile is a leading cause of nosocomial infections worldwide and has become an urgent public health threat requiring immediate attention. Epidemic lineages of the BI/NAP1/027 strain type have emerged and spread through health care systems across the globe over the past decade. Limiting person-to-person transmission and eradicating C. difficile, especially the BI/NAP1/027 strain type, from health care facilities are difficult due to the abundant shedding of spores that are impervious to most interventions. Effective prophylaxis for C. difficile infection (CDI) is lacking. We have genetically modified a contractile R-type bacteriocin ("diffocin") from C. difficile strain CD4 to kill BI/NAP1/027-type strains for this purpose. The natural receptor binding protein (RBP) responsible for diffocin targeting was replaced with a newly discovered RBP identified within a prophage of a BI/NAP1/027-type target strain by genome mining. The resulting modified diffocins (a.k.a. Avidocin-CDs), Av-CD291.1 and Av-CD291.2, were stable and killed all 16 tested BI/NAP1/027-type strains. Av-CD291.2 administered in drinking water survived passage through the mouse gastrointestinal (GI) tract, did not detectably alter the mouse gut microbiota or disrupt natural colonization resistance to C. difficile or the vancomycin-resistant Enterococcus faecium (VREF), and prevented antibiotic-induced colonization of mice inoculated with BI/NAP1/027-type spores. Given the high incidence and virulence of the pathogen, preventing colonization by BI/NAP1/027-type strains and limiting their transmission could significantly reduce the occurrence of the most severe CDIs. This modified diffocin represents a prototype of an Avidocin-CD platform capable of producing targetable, precision anti-C. difficile agents that can prevent and potentially treat CDIs without disrupting protective indigenous microbiota.
Importance: Treatment and prevention strategies for bacterial diseases rely heavily on traditional antibiotics, which impose strong selection for resistance and disrupt protective microbiota. One consequence has been an upsurge of opportunistic pathogens, such as Clostridium difficile, that exploit antibiotic-induced disruptions in gut microbiota to proliferate and cause life-threatening diseases. We have developed alternative agents that utilize contractile bactericidal protein complexes (R-type bacteriocins) to kill specific C. difficile pathogens. Efficacy in a preclinical animal study indicates these molecules warrant further development as potential prophylactic agents to prevent C. difficile infections in humans. Since these agents do not detectably alter the indigenous gut microbiota or colonization resistance in mice, we believe they will be safe to administer as a prophylactic to block transmission in high-risk environments without rendering patients susceptible to enteric infection after cessation of treatment.
Copyright © 2015 Gebhart et al.
Figures
Comment in
-
Antimicrobials: Targeting of C. difficile made easy.Nat Rev Microbiol. 2015 May;13(5):250. doi: 10.1038/nrmicro3481. Nat Rev Microbiol. 2015. PMID: 25895936 No abstract available.
References
-
- Centers for Disease Control and Prevention 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA.
-
- See I, Mu Y, Cohen J, Beldavs ZG, Winston LG, Dumyati G, Holzbauer S, Dunn J, Farley MM, Lyons C, Johnston H, Phipps E, Perlmutter R, Anderson L, Gerding DN, Lessa FC. 2014. NAP1 strain type predicts outcomes from Clostridium difficile infection. Clin Infect Dis 58:1394–1400. doi:10.1093/cid/ciu125. - DOI - PMC - PubMed
-
- Tickler IA, Goering RV, Whitmore JD, Lynn AN, Persing DH, Tenover FC, Healthcare Associated Infection Consortium . 2014. Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States: 2011–2013. Antimicrob Agents Chemother 58:4214–4218. doi:10.1128/AAC.02775-13. - DOI - PMC - PubMed
-
- He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D’Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi:10.1038/ng.2478. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

