StaRProtein, a web server for prediction of the stability of repeat proteins
- PMID: 25807112
- PMCID: PMC4373711
- DOI: 10.1371/journal.pone.0119417
StaRProtein, a web server for prediction of the stability of repeat proteins
Abstract
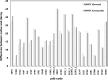
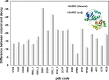
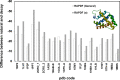
Repeat proteins have become increasingly important due to their capability to bind to almost any proteins and the potential as alternative therapy to monoclonal antibodies. In the past decade repeat proteins have been designed to mediate specific protein-protein interactions. The tetratricopeptide and ankyrin repeat proteins are two classes of helical repeat proteins that form different binding pockets to accommodate various partners. It is important to understand the factors that define folding and stability of repeat proteins in order to prioritize the most stable designed repeat proteins to further explore their potential binding affinities. Here we developed distance-dependant statistical potentials using two classes of alpha-helical repeat proteins, tetratricopeptide and ankyrin repeat proteins respectively, and evaluated their efficiency in predicting the stability of repeat proteins. We demonstrated that the repeat-specific statistical potentials based on these two classes of repeat proteins showed paramount accuracy compared with non-specific statistical potentials in: 1) discriminate correct vs. incorrect models 2) rank the stability of designed repeat proteins. In particular, the statistical scores correlate closely with the equilibrium unfolding free energies of repeat proteins and therefore would serve as a novel tool in quickly prioritizing the designed repeat proteins with high stability. StaRProtein web server was developed for predicting the stability of repeat proteins.
Conflict of interest statement
Figures

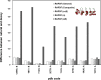
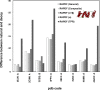


Similar articles
-
Ankyrin repeat: a unique motif mediating protein-protein interactions.Biochemistry. 2006 Dec 26;45(51):15168-78. doi: 10.1021/bi062188q. Biochemistry. 2006. PMID: 17176038 Review.
-
Consensus design as a tool for engineering repeat proteins.Methods Mol Biol. 2006;340:151-70. doi: 10.1385/1-59745-116-9:151. Methods Mol Biol. 2006. PMID: 16957336 Review.
-
Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins.J Mol Biol. 2008 Feb 8;376(1):241-57. doi: 10.1016/j.jmb.2007.11.046. Epub 2007 Nov 22. J Mol Biol. 2008. PMID: 18164721
-
The ankyrin repeat as molecular architecture for protein recognition.Protein Sci. 2004 Jun;13(6):1435-48. doi: 10.1110/ps.03554604. Protein Sci. 2004. PMID: 15152081 Free PMC article. Review.
-
A minimum folding unit in the ankyrin repeat protein p16(INK4).J Mol Biol. 2000 Jun 16;299(4):1121-32. doi: 10.1006/jmbi.2000.3803. J Mol Biol. 2000. PMID: 10843863
References
-
- Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J Struct Biol. 2001; 134:117–131. - PubMed
-
- Suzuki F, Goto M, Sawa C, Ito S, Watanabe H, Sawada J, et al. Functional interactions of transcription factor human GA-binding protein subunits. J Biol Chem. 1998; 273: 29302–29308. - PubMed
-
- Malek S, Huxford T & Ghosh G. IκBα functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF-κB. J Biol Chem. 1998; 273: 25427–25435. - PubMed
-
- Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins: Struct Funct Genet. 1993; 17: 363–374. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

