Acceleration of age-associated methylation patterns in HIV-1-infected adults
- PMID: 25807146
- PMCID: PMC4373843
- DOI: 10.1371/journal.pone.0119201
Acceleration of age-associated methylation patterns in HIV-1-infected adults
Abstract
Patients with treated HIV-1-infection experience earlier occurrence of aging-associated diseases, raising speculation that HIV-1-infection, or antiretroviral treatment, may accelerate aging. We recently described an age-related co-methylation module comprised of hundreds of CpGs; however, it is unknown whether aging and HIV-1-infection exert negative health effects through similar, or disparate, mechanisms. We investigated whether HIV-1-infection would induce age-associated methylation changes. We evaluated DNA methylation levels at >450,000 CpG sites in peripheral blood mononuclear cells (PBMC) of young (20-35) and older (36-56) adults in two separate groups of participants. Each age group for each data set consisted of 12 HIV-1-infected and 12 age-matched HIV-1-uninfected samples for a total of 96 samples. The effects of age and HIV-1 infection on methylation at each CpG revealed a strong correlation of 0.49, p<1 x 10(-200) and 0.47, p<1 x 10(-200). Weighted gene correlation network analysis (WGCNA) identified 17 co-methylation modules; module 3 (ME3) was significantly correlated with age (cor=0.70) and HIV-1 status (cor=0.31). Older HIV-1+ individuals had a greater number of hypermethylated CpGs across ME3 (p=0.015). In a multivariate model, ME3 was significantly associated with age and HIV status (Data set 1: βage=0.007088, p=2.08 x 10(-9); βHIV=0.099574, p=0.0011; Data set 2: βage=0.008762, p=1.27 x 10(-5); βHIV=0.128649, p=0.0001). Using this model, we estimate that HIV-1 infection accelerates age-related methylation by approximately 13.7 years in data set 1 and 14.7 years in data set 2. The genes related to CpGs in ME3 are enriched for polycomb group target genes known to be involved in cell renewal and aging. The overlap between ME3 and an aging methylation module found in solid tissues is also highly significant (Fisher-exact p=5.6 x 10(-6), odds ratio=1.91). These data demonstrate that HIV-1 infection is associated with methylation patterns that are similar to age-associated patterns and suggest that general aging and HIV-1 related aging work through some common cellular and molecular mechanisms. These results are an important first step for finding potential therapeutic targets and novel clinical approaches to mitigate the detrimental effects of both HIV-1-infection and aging.
Conflict of interest statement
Figures
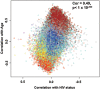

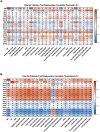
References
Publication types
MeSH terms
Grants and funding
- U01 AI035042/AI/NIAID NIH HHS/United States
- U01-AI35040/AI/NIAID NIH HHS/United States
- AI-28697/AI/NIAID NIH HHS/United States
- U01 AI035041/AI/NIAID NIH HHS/United States
- 5T32GM008243-25/GM/NIGMS NIH HHS/United States
- UL1 TR000424/TR/NCATS NIH HHS/United States
- U01-AI35041/AI/NIAID NIH HHS/United States
- U01-AI35039/AI/NIAID NIH HHS/United States
- UL1-TR000424/TR/NCATS NIH HHS/United States
- 5P30 AI028697/AI/NIAID NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
- UM1-AI35043/AI/NIAID NIH HHS/United States
- R01 AG030327/AG/NIA NIH HHS/United States
- U01-AI35042/AI/NIAID NIH HHS/United States
- P30 CA016042/CA/NCI NIH HHS/United States
- UM1 AI035043/AI/NIAID NIH HHS/United States
- T32 GM008243/GM/NIGMS NIH HHS/United States
- P30 AI028697/AI/NIAID NIH HHS/United States
- 1R01-AG-030327/AG/NIA NIH HHS/United States
- U01 AI035040/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

