Fast Volume Reconstruction From Motion Corrupted Stacks of 2D Slices
- PMID: 25807565
- PMCID: PMC7115883
- DOI: 10.1109/TMI.2015.2415453
Fast Volume Reconstruction From Motion Corrupted Stacks of 2D Slices
Abstract
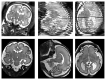
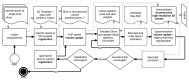
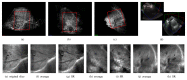
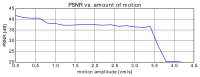
Capturing an enclosing volume of moving subjects and organs using fast individual image slice acquisition has shown promise in dealing with motion artefacts. Motion between slice acquisitions results in spatial inconsistencies that can be resolved by slice-to-volume reconstruction (SVR) methods to provide high quality 3D image data. Existing algorithms are, however, typically very slow, specialised to specific applications and rely on approximations, which impedes their potential clinical use. In this paper, we present a fast multi-GPU accelerated framework for slice-to-volume reconstruction. It is based on optimised 2D/3D registration, super-resolution with automatic outlier rejection and an additional (optional) intensity bias correction. We introduce a novel and fully automatic procedure for selecting the image stack with least motion to serve as an initial registration target. We evaluate the proposed method using artificial motion corrupted phantom data as well as clinical data, including tracked freehand ultrasound of the liver and fetal Magnetic Resonance Imaging. We achieve speed-up factors greater than 30 compared to a single CPU system and greater than 10 compared to currently available state-of-the-art multi-core CPU methods. We ensure high reconstruction accuracy by exact computation of the point-spread function for every input data point, which has not previously been possible due to computational limitations. Our framework and its implementation is scalable for available computational infrastructures and tests show a speed-up factor of 1.70 for each additional GPU. This paves the way for the online application of image based reconstruction methods during clinical examinations. The source code for the proposed approach is publicly available.
Figures



References
-
- Rohling R, Gee A, Berman L. Three-dimensional spatial compounding of ultrasound images. Medical Image Analysis. 1997;1(3):177–193. - PubMed
-
- Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza C, Vigneron DB, Barkovich JA, Studholme C. Registration-Based Approach for Reconstruction of High-Resolution In Utero Fetal MR Brain Images. Academic Radiology. 2006;13(9):1072–1081. - PubMed
-
- Karamalis A, Wein W, Kutter O, Navab N. Fast Hybrid Freehand Ultrasound Volume Reconstruction. In: Miga M, Wong I, Kenneth H, editors. Proc of the SPIE. Vol. 7261. 2009. pp. 726114–726118.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

