The SWI/SNF ATPases Are Required for Triple Negative Breast Cancer Cell Proliferation
- PMID: 25808524
- PMCID: PMC4516601
- DOI: 10.1002/jcp.24991
The SWI/SNF ATPases Are Required for Triple Negative Breast Cancer Cell Proliferation
Erratum in
-
Correction to "The SWI/SNF ATPases are required for triple negative breast cancer cell proliferation".J Cell Physiol. 2024 Nov;239(11):e31395. doi: 10.1002/jcp.31395. Epub 2024 Aug 27. J Cell Physiol. 2024. PMID: 39188216 No abstract available.
Abstract
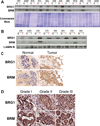
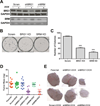
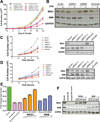
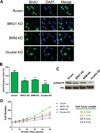
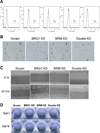
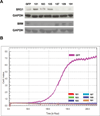
The Brahma (BRM) and Brahma-related Gene 1 (BRG1) ATPases are highly conserved homologs that catalyze the chromatin remodeling functions of the multi-subunit human SWI/SNF chromatin remodeling enzymes in a mutually exclusive manner. SWI/SNF enzyme subunits are mutated or missing in many cancer types, but are overexpressed without apparent mutation in other cancers. Here, we report that both BRG1 and BRM are overexpressed in most primary breast cancers independent of the tumor's receptor status. Knockdown of either ATPase in a triple negative breast cancer cell line reduced tumor formation in vivo and cell proliferation in vitro. Fewer cells in S phase and an extended cell cycle progression time were observed without any indication of apoptosis, senescence, or alterations in migration or attachment properties. Combined knockdown of BRM and BRG1 showed additive effects in the reduction of cell proliferation and time required for completion of cell cycle, suggesting that these enzymes promote cell cycle progression through independent mechanisms. Knockout of BRG1 or BRM using CRISPR/Cas9 technology resulted in the loss of viability, consistent with a requirement for both enzymes in triple negative breast cancer cells.
© 2015 Wiley Periodicals, Inc.
Figures
Similar articles
-
The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer.Epigenomics. 2017 Jun;9(6):919-931. doi: 10.2217/epi-2017-0034. Epub 2017 May 19. Epigenomics. 2017. PMID: 28521512 Free PMC article. Review.
-
The Bromodomains of the mammalian SWI/SNF (mSWI/SNF) ATPases Brahma (BRM) and Brahma Related Gene 1 (BRG1) promote chromatin interaction and are critical for skeletal muscle differentiation.Nucleic Acids Res. 2021 Aug 20;49(14):8060-8077. doi: 10.1093/nar/gkab617. Nucleic Acids Res. 2021. PMID: 34289068 Free PMC article.
-
SWI/SNF chromatin remodeling enzyme ATPases promote cell proliferation in normal mammary epithelial cells.J Cell Physiol. 2010 Jun;223(3):667-78. doi: 10.1002/jcp.22072. J Cell Physiol. 2010. PMID: 20333683 Free PMC article.
-
Co-regulation of transcription by BRG1 and BRM, two mutually exclusive SWI/SNF ATPase subunits.Epigenetics Chromatin. 2017 Dec 22;10(1):62. doi: 10.1186/s13072-017-0167-8. Epigenetics Chromatin. 2017. PMID: 29273066 Free PMC article.
-
Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer.Epigenetics. 2008 Mar-Apr;3(2):64-8. doi: 10.4161/epi.3.2.6153. Epub 2008 Apr 17. Epigenetics. 2008. PMID: 18437052 Review.
Cited by
-
Brg1 controls stemness and metastasis of pancreatic cancer through regulating hypoxia pathway.Oncogene. 2023 Jun;42(26):2139-2152. doi: 10.1038/s41388-023-02716-4. Epub 2023 May 18. Oncogene. 2023. PMID: 37198398
-
SNF5 promotes cell proliferation and immune evasion in non-small cell lung cancer.Bioengineered. 2022 May;13(5):11530-11540. doi: 10.1080/21655979.2022.2068894. Bioengineered. 2022. PMID: 35506290 Free PMC article.
-
Identification of key genes involved in tamoxifen-resistant breast cancer using bioinformatics analysis.Transl Cancer Res. 2021 Dec;10(12):5246-5257. doi: 10.21037/tcr-21-1276. Transl Cancer Res. 2021. PMID: 35116374 Free PMC article.
-
High expression of SMARCA4 or SMARCA2 is frequently associated with an opposite prognosis in cancer.Sci Rep. 2018 Feb 1;8(1):2043. doi: 10.1038/s41598-018-20217-3. Sci Rep. 2018. PMID: 29391527 Free PMC article.
-
The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer.Epigenomics. 2017 Jun;9(6):919-931. doi: 10.2217/epi-2017-0034. Epub 2017 May 19. Epigenomics. 2017. PMID: 28521512 Free PMC article. Review.
References
-
- Bishop JM. Enemies within: the genesis of retrovirus oncogenes. Cell. 1981;23(1):5–6. - PubMed
-
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102(2):257–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

