Identification and characterization of diverse groups of endogenous retroviruses in felids
- PMID: 25808580
- PMCID: PMC4373062
- DOI: 10.1186/s12977-015-0152-x
Identification and characterization of diverse groups of endogenous retroviruses in felids
Abstract
Background: Endogenous retroviruses (ERVs) are genetic elements with a retroviral origin that are integrated into vertebrate genomes. In felids (Mammalia, Carnivora, Felidae), ERVs have been described mostly in the domestic cat, and only rarely in wild species. To gain insight into the origins and evolutionary dynamics of endogenous retroviruses in felids, we have identified and characterized partial pro/pol ERV sequences from eight Neotropical wild cat species, belonging to three distinct lineages of Felidae. We also compared them with publicly available genomic sequences of Felis catus and Panthera tigris, as well as with representatives of other vertebrate groups, and performed phylogenetic and molecular dating analyses to investigate the pattern and timing of diversification of these retroviral elements.
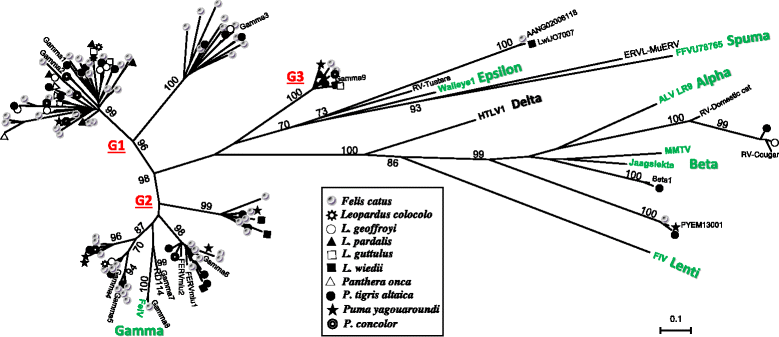
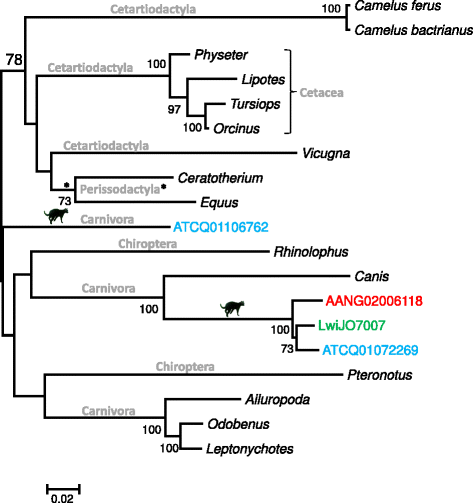
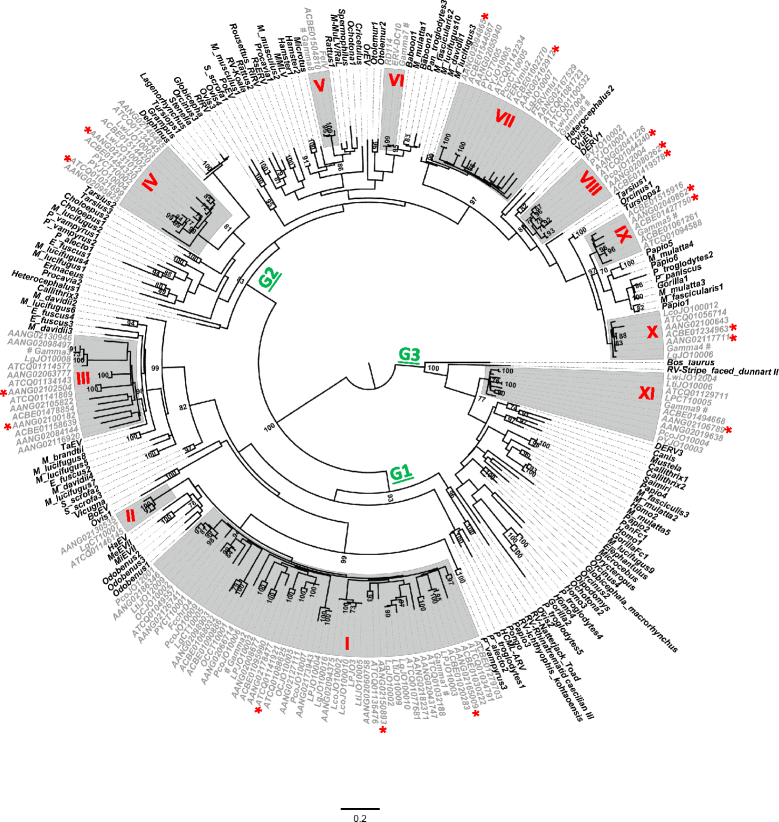
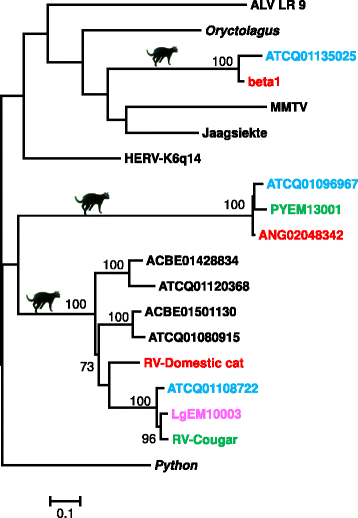
Results: We identified a high diversity of ERVs in the sampled felids, with a predominance of Gammaretrovirus-related sequences, including class I ERVs. Our data indicate that the identified ERVs arose from at least eleven horizontal interordinal transmissions from other mammals. Furthermore, we estimated that the majority of the Gamma-like integrations took place during the diversification of modern felids. Finally, our phylogenetic analyses indicate the presence of a genetically divergent group of sequences whose position in our phylogenetic tree was difficult to establish confidently relative to known retroviruses, and another lineage identified as ERVs belonging to class II.
Conclusions: Retroviruses have circulated in felids along with their evolution. The majority of the deep clades of ERVs exist since the primary divergence of felids' base and cluster with retroviruses of divergent mammalian lineages, suggesting horizontal interordinal transmission. Our findings highlight the importance of additional studies on the role of ERVs in the genome landscaping of other carnivore species.
Figures
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

