Mitochondrial protein import receptors in Kinetoplastids reveal convergent evolution over large phylogenetic distances
- PMID: 25808593
- PMCID: PMC4389251
- DOI: 10.1038/ncomms7646
Mitochondrial protein import receptors in Kinetoplastids reveal convergent evolution over large phylogenetic distances
Abstract
Mitochondrial protein import is essential for all eukaryotes and mediated by hetero-oligomeric protein translocases thought to be conserved within all eukaryotes. We have identified and analysed the function and architecture of the non-conventional outer membrane (OM) protein translocase in the early diverging eukaryote Trypanosoma brucei. It consists of six subunits that show no obvious homology to translocase components of other species. Two subunits are import receptors that have a unique topology and unique protein domains and thus evolved independently of the prototype receptors Tom20 and Tom70. Our study suggests that protein import receptors were recruited to the core of the OM translocase after the divergence of the major eukaryotic supergroups. Moreover, it links the evolutionary history of mitochondrial protein import receptors to the origin of the eukaryotic supergroups.
Figures
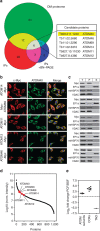

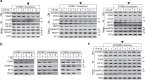
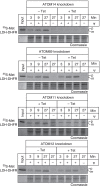




References
-
- Dolezal P., Likic V., Tachezy J. & Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 (2006) . - PubMed
-
- Hewitt V., Alcock F. & Lithgow T. Minor modifications and major adaptations: the evolution of molecular machines driving mitochondrial protein import. Biochim. Biophys. Acta 1808, 947–954 (2011) . - PubMed
-
- Schmidt O., Pfanner N. & Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 (2010) . - PubMed
-
- Neupert W. & Herrmann J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 (2007) . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

