Stat5 Exerts Distinct, Vital Functions in the Cytoplasm and Nucleus of Bcr-Abl+ K562 and Jak2(V617F)+ HEL Leukemia Cells
- PMID: 25809097
- PMCID: PMC4381271
- DOI: 10.3390/cancers7010503
Stat5 Exerts Distinct, Vital Functions in the Cytoplasm and Nucleus of Bcr-Abl+ K562 and Jak2(V617F)+ HEL Leukemia Cells
Erratum in
-
Correction: Weber et al. Stat5 Exerts Distinct, Vital Functions in the Cytoplasm and Nucleus of Bcr-Abl+ K562 and Jak2(V617F)+ HEL Leukemia Cells. Cancers 2015, 7, 503-537.Cancers (Basel). 2025 May 23;17(11):1754. doi: 10.3390/cancers17111754. Cancers (Basel). 2025. PMID: 40445629 Free PMC article.
Abstract
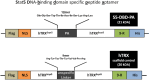
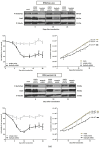
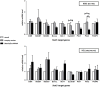
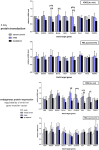
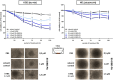
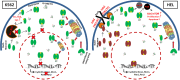
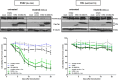
Signal transducers and activators of transcription (Stats) play central roles in the conversion of extracellular signals, e.g., cytokines, hormones and growth factors, into tissue and cell type specific gene expression patterns. In normal cells, their signaling potential is strictly limited in extent and duration. The persistent activation of Stat3 or Stat5 is found in many human tumor cells and contributes to their growth and survival. Stat5 activation plays a pivotal role in nearly all hematological malignancies and occurs downstream of oncogenic kinases, e.g., Bcr-Abl in chronic myeloid leukemias (CML) and Jak2(V617F) in other myeloproliferative diseases (MPD). We defined the mechanisms through which Stat5 affects growth and survival of K562 cells, representative of Bcr-Abl positive CML, and HEL cells, representative for Jak2(V617F) positive acute erythroid leukemia. In our experiments we suppressed the protein expression levels of Stat5a and Stat5b through shRNA mediated downregulation and demonstrated the dependence of cell survival on the presence of Stat5. Alternatively, we interfered with the functional capacities of the Stat5 protein through the interaction with a Stat5 specific peptide ligand. This ligand is a Stat5 specific peptide aptamer construct which comprises a 12mer peptide integrated into a modified thioredoxin scaffold, S5-DBD-PA. The peptide sequence specifically recognizes the DNA binding domain (DBD) of Stat5. Complex formation of S5-DBD-PA with Stat5 causes a strong reduction of P-Stat5 in the nuclear fraction of Bcr-Abl-transformed K562 cells and a suppression of Stat5 target genes. Distinct Stat5 mediated survival mechanisms were detected in K562 and Jak2(V617F)-transformed HEL cells. Stat5 is activated in the nuclear and cytosolic compartments of K562 cells and the S5-DBD-PA inhibitor most likely affects the viability of Bcr-Abl+ K562 cells through the inhibition of canonical Stat5 induced target gene transcription. In HEL cells, Stat5 is predominantly present in the cytoplasm and the survival of the Jak2(V617F)+ HEL cells is impeded through the inhibition of the cytoplasmic functions of Stat5.
Figures









References
-
- Groner B., Hennighausen L. The versatile regulation of cellular events by jak-stat signaling: From transcriptional control to microtubule dynamics and energy metabolism. Horm. Mol. Biol. Clin. Investg. 2012;10:193–200. - PubMed
-
- Vafaizadeh V., Klemmt P., Brendel C., Weber K., Doebele C., Britt K., Grez M., Fehse B., Desrivieres S., Groner B. Mammary epithelial reconstitution with gene-modified stem cells assigns roles to stat5 in luminal alveolar cell fate decisions, differentiation, involution, and mammary tumor formation. Stem Cells. 2010;28:928–938. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

