Probing the structural and dynamical effects of the charged residues of the TZF domain of TIS11d
- PMID: 25809263
- PMCID: PMC4375430
- DOI: 10.1016/j.bpj.2015.01.039
Probing the structural and dynamical effects of the charged residues of the TZF domain of TIS11d
Abstract
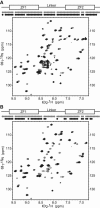
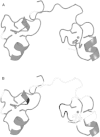
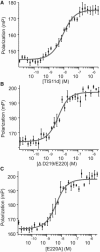
A member of the TTP family of proteins, TIS11d binds RNA with high specificity using a pair of CCCH-type tandem zinc fingers separated by a 18 residue long linker. Our previous work showed that the formation of hydrogen bonds between the C-terminal residue E220 and the residues of the linker region stabilized a compact structure of TIS11d in the absence of RNA. To investigate the role of the C-terminal residues in the structure of unbound TIS11d, the E220A mutant and the truncation mutant lacking the last two residues (D219/E220) were studied using molecular dynamics, NMR spectroscopy, and biochemical methods. This study confirmed the importance of the charged residues D219 and E220 in maintaining structural stability in unbound TIS11d and elucidated the underlying physical mechanisms. We observed a greater structural heterogeneity for the residues of the linker in the molecular dynamics trajectories of both mutant proteins relative to the wild-type. This heterogeneity was more pronounced in the D219/E220 deletion mutant than in the E220A mutant, indicating that a greater reduction of the charge of the C-terminus results in greater flexibility. In agreement with the increased flexibility and the reduced number of negatively charged residues of the D219/E220 deletion mutant, we measured more unfavorable entropic and a more favorable enthalpic contribution to the free energy of RNA binding in the mutant than in the wild-type protein. The relative orientation of the zinc fingers was stabilized by the electrostatic interaction between E220 and positively charged residues of the linker in TIS11d. In the E220A mutant, the relative orientation of the zinc fingers was less constrained, whereas in the D219/E220 deletion mutant, little orientational preference was observed. We posit that favorable electrostatic interactions provide a mechanism to promote preferential orientation of separate domains without imposing structural rigidity.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Carballo E., Lai W.S., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. - PubMed
-
- Chen C.Y., Gherzi R., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. - PubMed
-
- Blackshear P.J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. - PubMed
-
- Brewer B.Y., Malicka J., Wilson G.M. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J. Biol. Chem. 2004;279:27870–27877. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

