Phosphorylation of Minichromosome Maintenance 3 (MCM3) by Checkpoint Kinase 1 (Chk1) Negatively Regulates DNA Replication and Checkpoint Activation
- PMID: 25809478
- PMCID: PMC4424366
- DOI: 10.1074/jbc.M114.621532
Phosphorylation of Minichromosome Maintenance 3 (MCM3) by Checkpoint Kinase 1 (Chk1) Negatively Regulates DNA Replication and Checkpoint Activation
Abstract
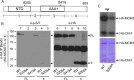
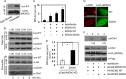
Mechanisms controlling DNA replication and replication checkpoint are critical for the maintenance of genome stability and the prevention or treatment of human cancers. Checkpoint kinase 1 (Chk1) is a key effector protein kinase that regulates the DNA damage response and replication checkpoint. The heterohexameric minichromosome maintenance (MCM) complex is the core component of mammalian DNA helicase and has been implicated in replication checkpoint activation. Here we report that Chk1 phosphorylates the MCM3 subunit of the MCM complex at Ser-205 under normal growth conditions. Mutating the Ser-205 of MCM3 to Ala increased the length of DNA replication track and shortened the S phase duration, indicating that Ser-205 phosphorylation negatively controls normal DNA replication. Upon replicative stress treatment, the inhibitory phosphorylation of MCM3 at Ser-205 was reduced, and this reduction was accompanied with the generation of single strand DNA, the key platform for ataxia telangiectasia mutated and Rad3-related (ATR) activation. As a result, the replication checkpoint is activated. Together, these data provide significant insights into the regulation of both normal DNA replication and replication checkpoint activation through the novel phosphorylation of MCM3 by Chk1.
Keywords: Cell Cycle; DNA Damage; DNA Replication; Phosphorylation; Signal Transduction.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
The interaction between checkpoint kinase 1 (Chk1) and the minichromosome maintenance (MCM) complex is required for DNA damage-induced Chk1 phosphorylation.J Biol Chem. 2014 Aug 29;289(35):24716-23. doi: 10.1074/jbc.M114.575035. Epub 2014 Jul 21. J Biol Chem. 2014. PMID: 25049228 Free PMC article.
-
DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response.Nucleic Acids Res. 2014 Apr;42(7):4463-73. doi: 10.1093/nar/gku116. Epub 2014 Feb 5. Nucleic Acids Res. 2014. PMID: 24500207 Free PMC article.
-
Identification of carboxyl-terminal MCM3 phosphorylation sites using polyreactive phosphospecific antibodies.J Biol Chem. 2007 Mar 23;282(12):9236-43. doi: 10.1074/jbc.M609256200. Epub 2007 Jan 23. J Biol Chem. 2007. PMID: 17244605
-
Regulation of ATR-CHK1 signaling by ubiquitination of CLASPIN.Biochem Soc Trans. 2022 Oct 31;50(5):1471-1480. doi: 10.1042/BST20220729. Biochem Soc Trans. 2022. PMID: 36196914 Review.
-
The Hammer and the Dance of Cell Cycle Control.Trends Biochem Sci. 2021 Apr;46(4):301-314. doi: 10.1016/j.tibs.2020.11.002. Epub 2020 Dec 2. Trends Biochem Sci. 2021. PMID: 33279370 Review.
Cited by
-
Diagnostic and prognostic value of MCM3 and its interacting proteins in hepatocellular carcinoma.Oncol Lett. 2020 Dec;20(6):308. doi: 10.3892/ol.2020.12171. Epub 2020 Sep 30. Oncol Lett. 2020. PMID: 33093917 Free PMC article.
-
Impact of Age and Insulin-Like Growth Factor-1 on DNA Damage Responses in UV-Irradiated Human Skin.Molecules. 2017 Feb 26;22(3):356. doi: 10.3390/molecules22030356. Molecules. 2017. PMID: 28245638 Free PMC article. Review.
-
ZMIZ2 promotes the development of triple-receptor negative breast cancer.Cancer Cell Int. 2022 Jan 31;22(1):52. doi: 10.1186/s12935-021-02393-x. Cancer Cell Int. 2022. PMID: 35101047 Free PMC article.
-
On the Interplay of the DNA Replication Program and the Intra-S Phase Checkpoint Pathway.Genes (Basel). 2019 Jan 29;10(2):94. doi: 10.3390/genes10020094. Genes (Basel). 2019. PMID: 30700024 Free PMC article. Review.
-
Tyrosyl-DNA phosphodiesterases are involved in mutagenic events at a ribonucleotide embedded into DNA in human cells.PLoS One. 2020 Dec 31;15(12):e0244790. doi: 10.1371/journal.pone.0244790. eCollection 2020. PLoS One. 2020. PMID: 33382846 Free PMC article.
References
-
- Harper J. W., Elledge S. J. (2007) The DNA damage response: ten years after. Mol. Cell 28, 739–745 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

