Self-cleavage of the Pseudomonas aeruginosa Cell-surface Signaling Anti-sigma Factor FoxR Occurs through an N-O Acyl Rearrangement
- PMID: 25809487
- PMCID: PMC4424355
- DOI: 10.1074/jbc.M115.643098
Self-cleavage of the Pseudomonas aeruginosa Cell-surface Signaling Anti-sigma Factor FoxR Occurs through an N-O Acyl Rearrangement
Abstract
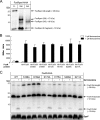
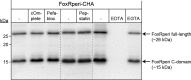
The Fox system of Pseudomonas aeruginosa is a cell-surface signaling (CSS) pathway employed by the bacterium to sense and respond to the presence of the heterologous siderophore ferrioxamine in the environment. This regulatory pathway controls the transcription of the foxA ferrioxamine receptor gene through the extracytoplasmic function sigma factor σ(FoxI). In the absence of ferrioxamine, the activity of σ(FoxI) is inhibited by the transmembrane anti-sigma factor FoxR. Upon binding of ferrioxamine by the FoxA receptor, FoxR is processed by a complex proteolytic cascade leading to the release and activation of σ(FoxI). Interestingly, we have recently shown that FoxR undergoes self-cleavage between the periplasmic Gly-191 and Thr-192 residues independent of the perception of ferrioxamine. This autoproteolytic event, which is widespread among CSS anti-sigma factors, produces two distinct domains that interact and function together to transduce the presence of the signal. In this work, we provide evidence that the self-cleavage of FoxR is not an enzyme-dependent process but is induced by an N-O acyl rearrangement. Mutation analysis showed that the nucleophilic side chain of the Thr-192 residue at +1 of the cleavage site is required for an attack on the preceding Gly-191, after which the resulting ester bond is likely hydrolyzed. Because the cleavage site is well preserved and the hydrolysis of periplasmic CSS anti-sigma factors is widely observed, we hypothesize that cleavage via an N-O acyl rearrangement is a conserved feature of these proteins.
Keywords: Cell Signaling; Gene Regulation; Iron; Post-translational Modification (PTM); Proteolysis; Pseudomonas aeruginosa (P. aeruginosa); Siderophore; Signal Transduction.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Ratledge C., Dover L. G. (2000) Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941 - PubMed
-
- Wandersman C., Delepelaire P. (2004) Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58, 611–647 - PubMed
-
- Poole K., McKay G. A. (2003) Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8, d661–d686 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

