The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis
- PMID: 25810095
- PMCID: PMC4424013
- DOI: 10.1104/pp.114.255000
The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis
Abstract
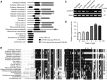
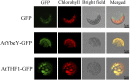
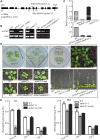
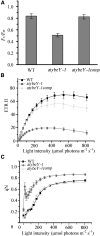
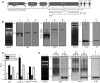
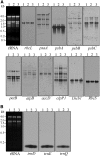
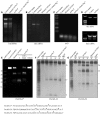
Maturation of chloroplast ribosomal RNAs (rRNAs) comprises several endoribonucleolytic and exoribonucleolytic processing steps. However, little is known about the specific enzymes involved and the cleavage steps they catalyze. Here, we report the functional characterization of the single Arabidopsis (Arabidopsis thaliana) gene encoding a putative YbeY endoribonuclease. AtYbeY null mutants are seedling lethal, indicating that AtYbeY function is essential for plant growth. Knockdown plants display slow growth and show pale-green leaves. Physiological and ultrastructural analyses of atybeY mutants revealed impaired photosynthesis and defective chloroplast development. Fluorescent microcopy analysis showed that, when fused with the green fluorescence protein, AtYbeY is localized in chloroplasts. Immunoblot and RNA gel-blot assays revealed that the levels of chloroplast-encoded subunits of photosynthetic complexes are reduced in atybeY mutants, but the corresponding transcripts accumulate normally. In addition, atybeY mutants display defective maturation of both the 5' and 3' ends of 16S, 23S, and 4.5S rRNAs as well as decreased accumulation of mature transcripts from the transfer RNA genes contained in the chloroplast rRNA operon. Consequently, mutant plants show a severe deficiency in ribosome biogenesis, which, in turn, results in impaired plastid translational activity. Furthermore, biochemical assays show that recombinant AtYbeY is able to cleave chloroplast rRNAs as well as messenger RNAs and transfer RNAs in vitro. Taken together, our findings indicate that AtYbeY is a chloroplast-localized endoribonuclease that is required for chloroplast rRNA processing and thus for normal growth and development.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Allen JF, Forsberg J (2001) Molecular recognition in thylakoid structure and function. Trends Plant Sci 6: 317–326 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Bang WY, Chen J, Jeong IS, Kim SW, Kim CW, Jung HS, Lee KH, Kweon HS, Yoko I, Shiina T, et al. (2012) Functional characterization of ObgC in ribosome biogenesis during chloroplast development. Plant J 71: 122–134 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

