S-sulfhydration: a cysteine posttranslational modification in plant systems
- PMID: 25810097
- PMCID: PMC4424021
- DOI: 10.1104/pp.15.00009
S-sulfhydration: a cysteine posttranslational modification in plant systems
Abstract
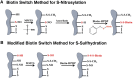
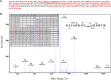
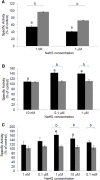
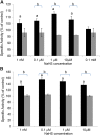
Hydrogen sulfide is a highly reactive molecule that is currently accepted as a signaling compound. This molecule is as important as carbon monoxide in mammals and hydrogen peroxide in plants, as well as nitric oxide in both eukaryotic systems. Although many studies have been conducted on the physiological effects of hydrogen sulfide, the underlying mechanisms are poorly understood. One of the proposed mechanisms involves the posttranslational modification of protein cysteine residues, a process called S-sulfhydration. In this work, a modified biotin switch method was used for the detection of Arabidopsis (Arabidopsis thaliana) proteins modified by S-sulfhydration under physiological conditions. The presence of an S-sulfhydration-modified cysteine residue on cytosolic ascorbate peroxidase was demonstrated using liquid chromatography-tandem mass spectrometry analysis, and a total of 106 S-sulfhydrated proteins were identified. Immunoblot and enzyme activity analyses of some of these proteins showed that the sulfide added through S-sulfhydration reversibly regulates the functions of plant proteins in a manner similar to that described in mammalian systems.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Álvarez C, Bermúdez MA, Romero LC, Gotor C, García I (2012a) Cysteine homeostasis plays an essential role in plant immunity. New Phytol 193: 165–177 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

