Polyhydroxylated [60]fullerene binds specifically to functional recognition sites on a monomeric and a dimeric ubiquitin
- PMID: 25811293
- PMCID: PMC4443925
- DOI: 10.1039/c5nr00539f
Polyhydroxylated [60]fullerene binds specifically to functional recognition sites on a monomeric and a dimeric ubiquitin
Abstract
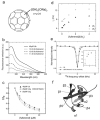
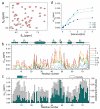
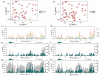
The use of nanoparticles (NPs) in biomedical applications requires an in-depth understanding of the mechanisms by which NPs interact with biomolecules. NPs associating with proteins may interfere with protein-protein interactions and affect cellular communication pathways, however the impact of NPs on biomolecular recognition remains poorly characterized. In this respect, particularly relevant is the study of NP-induced functional perturbations of proteins implicated in the regulation of key biochemical pathways. Ubiquitin (Ub) is a prototypical protein post-translational modifier playing a central role in numerous essential biological processes. To contribute to the understanding of the interactions between this universally distributed biomacromolecule and NPs, we investigated the adsorption of polyhydroxylated [60]fullerene on monomeric Ub and on a minimal polyubiquitin chain in vitro at atomic resolution. Site-resolved chemical shift and intensity perturbations of Ub's NMR signals, together with (15)N spin relaxation rate changes, exchange saturation transfer effects, and fluorescence quenching data were consistent with the reversible formation of soluble aggregates incorporating fullerenol clusters. The specific interaction epitopes were identified, coincident with functional recognition sites in a monomeric and lysine48-linked dimeric Ub. Fullerenol appeared to target the open state of the dynamic structure of a dimeric Ub according to a conformational selection mechanism. Importantly, the protein-NP association prevented the enzyme-catalyzed synthesis of polyubiquitin chains. Our findings provide an experiment-based insight into protein/fullerenol recognition, with implications in functional biomolecular communication, including regulatory protein turnover, and for the opportunity of therapeutic intervention in Ub-dependent cellular pathways.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

