Signature wood modifications reveal decomposer community history
- PMID: 25811364
- PMCID: PMC4374725
- DOI: 10.1371/journal.pone.0120679
Signature wood modifications reveal decomposer community history
Erratum in
-
Correction: Signature wood modifications reveal decomposer community history.PLoS One. 2015 Apr 23;10(4):e0126877. doi: 10.1371/journal.pone.0126877. eCollection 2015. PLoS One. 2015. PMID: 25906351 Free PMC article. No abstract available.
Abstract
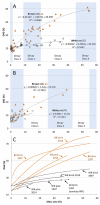
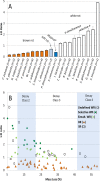
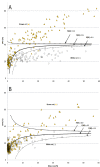
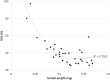
Correlating plant litter decay rates with initial tissue traits (e.g. C, N contents) is common practice, but in woody litter, predictive relationships are often weak. Variability in predicting wood decomposition is partially due to territorial competition among fungal decomposers that, in turn, have a range of nutritional strategies (rot types) and consequences on residues. Given this biotic influence, researchers are increasingly using culture-independent tools in an attempt to link variability more directly to decomposer groups. Our goal was to complement these tools by using certain wood modifications as 'signatures' that provide more functional information about decomposer dominance than density loss. Specifically, we used dilute alkali solubility (DAS; higher for brown rot) and lignin:density loss (L:D; higher for white rot) to infer rot type (binary) and fungal nutritional mode (gradient), respectively. We first determined strength of pattern among 29 fungi of known rot type by correlating DAS and L:D with mass loss in birch and pine. Having shown robust relationships for both techniques above a density loss threshold, we then demonstrated and resolved two issues relevant to species consortia and field trials, 1) spatial patchiness creating gravimetric bias (density bias), and 2) brown rot imprints prior or subsequent to white rot replacement (legacy effects). Finally, we field-tested our methods in a New Zealand Pinus radiata plantation in a paired-plot comparison. Overall, results validate these low-cost techniques that measure the collective histories of decomposer dominance in wood. The L:D measure also showed clear potential in classifying 'rot type' along a spectrum rather than as a traditional binary type (brown versus white rot), as it places the nutritional strategies of wood-degrading fungi on a scale (L:D=0-5, in this case). These information-rich measures of consequence can provide insight into their biological causes, strengthening the links between traits, structure, and function during wood decomposition.
Conflict of interest statement
Figures

References
-
- Eriksson KEL, Blanchette RA, Ander P (1990) Microbial and enzymatic degradation of wood and wood components. Berlin: Springer-Verlag; 407 p.
-
- Zabel RA, Morrell JJ (1992) Wood microbiology: decay and its prevention. San Diego: Academic Press, Inc; 476 p.
-
- Worrall JJ, Anagnost SE, Zabel RA (1997) Comparison of wood decay among diverse lignicolous fungi. Mycologia 89: 199–219.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

