Clinical outcome and genetic differences within a monophyletic Dengue virus type 2 population
- PMID: 25811657
- PMCID: PMC4374945
- DOI: 10.1371/journal.pone.0121696
Clinical outcome and genetic differences within a monophyletic Dengue virus type 2 population
Abstract
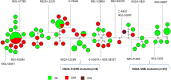
The exact mechanisms of interplay between host and viral factors leading to severe dengue are yet to be fully understood. Even though previous studies have implicated specific genetic differences of Dengue virus (DENV) in clinical severity and virus attenuation, similar studies with large-scale, whole genome screening of monophyletic virus populations are limited. Therefore, in the present study, we compared 89 whole genomes of DENV-2 cosmopolitan clade III isolates obtained from patients diagnosed with dengue fever (DF, n = 58), dengue hemorrhagic fever (DHF, n = 30) and dengue shock syndrome (DSS, n = 1) in Singapore between July 2010 and January 2013, in order to determine the correlation of observed viral genetic differences with clinical outcomes. Our findings showed no significant difference between the number of primary and secondary infections that progressed to DHF and DSS (p>0.05) in our study cohort. Despite being highly homogenous, study isolates possessed 39 amino acid substitutions of which 10 substitutions were fixed in three main groups of virus isolates. None of those substitutions were specifically associated with DHF and DSS. Notably, two evolutionarily unique virus groups possessing C-P43T+NS1-S103T+NS2A-V83I+NS3-R337K+ NS3-I600T+ NS5-P136S and NS2A-T119N mutations were exclusively found in patients with DF, the benign form of DENV infections. Those mutants were significantly associated with mild disease outcome. These observations indicated that disease progression into DHF and DSS within our patient population was more likely to be due to host than virus factors. We hypothesize that selection for potentially less virulent groups of DENV-2 in our study cohort may be an evolutionary adaptation of viral strains to extend their survival in the human-mosquito transmission cycle.
Conflict of interest statement
Figures



References
-
- WHO. Dengue: Guidelines for diagnosis, treatment, prevention and control New ed. Geneva: World Health Organization; 2009. - PubMed
-
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–88. - PubMed
-
- Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000. Feb;8(2):74–7. - PubMed
-
- Holmes EC, Twiddy SS. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol. 2003. May;3(1):19–28. - PubMed
-
- Twiddy SS, Woelk CH, Holmes EC. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J Gen Virol. 2002. Jul;83(Pt 7):1679–89. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

