The in vivo dynamics of antigenic variation in Trypanosoma brucei
- PMID: 25814582
- PMCID: PMC4514441
- DOI: 10.1126/science.aaa4502
The in vivo dynamics of antigenic variation in Trypanosoma brucei
Abstract
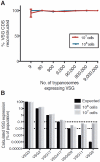
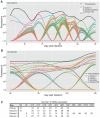
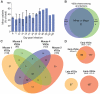
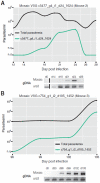
Trypanosoma brucei, a causative agent of African Sleeping Sickness, constantly changes its dense variant surface glycoprotein (VSG) coat to avoid elimination by the immune system of its mammalian host, using an extensive repertoire of dedicated genes. However, the dynamics of VSG expression in T. brucei during an infection are poorly understood. We have developed a method, based on de novo assembly of VSGs, for quantitatively examining the diversity of expressed VSGs in any population of trypanosomes and monitored VSG population dynamics in vivo. Our experiments revealed unexpected diversity within parasite populations and a mechanism for diversifying the genome-encoded VSG repertoire. The interaction between T. brucei and its host is substantially more dynamic and nuanced than previously expected.
Copyright © 2015, American Association for the Advancement of Science.
Figures
References
-
- Cross GAM, Kim HS, Wickstead B. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol Biochem Parasitol. 2014;195:59–73. - PubMed
-
- Ross R, Thomson D. A Case of Sleeping Sickness Studied by Precise Enumerative Methods: Regular Periodical Increase of the Parasites Disclosed. Proceedings of the Royal Society B: Biological Sciences. 1910;82:411–415.
-
- Turner CM, Barry JD. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology. 1989;99(Pt 1):67–75. - PubMed
-
- Turner CM. The rate of antigenic variation in fly-transmitted and syringe-passaged infections of Trypanosoma brucei. FEMS Microbiol Lett. 1997;153:227–231. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

