Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma
- PMID: 25814782
- PMCID: PMC4357047
- DOI: 10.1155/2015/893594
Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma
Abstract
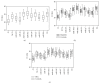
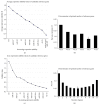
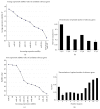
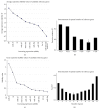
Serum exosomal microRNAs (miRNAs) have received considerable attention as potential biomarkers for diagnosing cancer. The canonical technique for measuring miRNA transcript levels is reverse transcription quantitative polymerase chain reaction (RT-qPCR). One prerequisite for validating RT-qPCR data is proper normalization with respect to stably expressed endogenous reference genes. However, genes that meet all of the criteria of a control gene for exosomal miRNAs have not yet been identified. To find out the control gene for exosomal miRNAs, we evaluated the expression stability of 11 well-known reference genes in circulating exosomes. In this study, we found that the combination of miR-221, miR-191, let-7a, miR-181a, and miR-26a can be an optimal gene reference set for normalizing the expression of liver-specific miRNAs. This combination enhanced the robustness of the relative quantification analyses. These findings highlight the importance of validating reference genes before quantifying target miRNAs. Furthermore, our findings will improve studies that monitor hepatitis progression and will aid in the discovery of noninvasive biomarkers to diagnose early stage HCC.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

