Biomarker-based classification of bacterial and fungal whole-blood infections in a genome-wide expression study
- PMID: 25814982
- PMCID: PMC4356159
- DOI: 10.3389/fmicb.2015.00171
Biomarker-based classification of bacterial and fungal whole-blood infections in a genome-wide expression study
Abstract
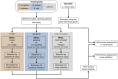
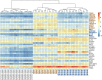
Sepsis is a clinical syndrome that can be caused by bacteria or fungi. Early knowledge on the nature of the causative agent is a prerequisite for targeted anti-microbial therapy. Besides currently used detection methods like blood culture and PCR-based assays, the analysis of the transcriptional response of the host to infecting organisms holds great promise. In this study, we aim to examine the transcriptional footprint of infections caused by the bacterial pathogens Staphylococcus aureus and Escherichia coli and the fungal pathogens Candida albicans and Aspergillus fumigatus in a human whole-blood model. Moreover, we use the expression information to build a random forest classifier to classify if a sample contains a bacterial, fungal, or mock-infection. After normalizing the transcription intensities using stably expressed reference genes, we filtered the gene set for biomarkers of bacterial or fungal blood infections. This selection is based on differential expression and an additional gene relevance measure. In this way, we identified 38 biomarker genes, including IL6, SOCS3, and IRG1 which were already associated to sepsis by other studies. Using these genes, we trained the classifier and assessed its performance. It yielded a 96% accuracy (sensitivities >93%, specificities >97%) for a 10-fold stratified cross-validation and a 92% accuracy (sensitivities and specificities >83%) for an additional test dataset comprising Cryptococcus neoformans infections. Furthermore, the classifier is robust to Gaussian noise, indicating correct class predictions on datasets of new species. In conclusion, this genome-wide approach demonstrates an effective feature selection process in combination with the construction of a well-performing classification model. Further analyses of genes with pathogen-dependent expression patterns can provide insights into the systemic host responses, which may lead to new anti-microbial therapeutic advances.
Keywords: decision tree based methods; feature selection; fungal pathogens; immune response; microarray; systems biology.
Figures


References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soci. Ser. B 57, 289–300.
-
- Breiman L. (2001). Random forests. Mach. Learn. 45, 5–32 10.1023/A:1010933404324 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

