H-ferritin-regulated microRNAs modulate gene expression in K562 cells
- PMID: 25815883
- PMCID: PMC4376865
- DOI: 10.1371/journal.pone.0122105
H-ferritin-regulated microRNAs modulate gene expression in K562 cells
Abstract
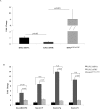
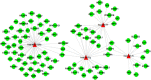
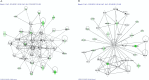
In a previous study, we showed that the silencing of the heavy subunit (FHC) offerritin, the central iron storage molecule in the cell, is accompanied by a modification in global gene expression. In this work, we explored whether different FHC amounts might modulate miRNA expression levels in K562 cells and studied the impact of miRNAs in gene expression profile modifications. To this aim, we performed a miRNA-mRNA integrative analysis in K562 silenced for FHC (K562shFHC) comparing it with K562 transduced with scrambled RNA (K562shRNA). Four miRNAs, namely hsa-let-7g, hsa-let-7f, hsa-let-7i and hsa-miR-125b, were significantly up-regulated in silenced cells. The remarkable down-regulation of these miRNAs, following FHC expression rescue, supports a specific relation between FHC silencing and miRNA-modulation. The integration of target predictions with miRNA and gene expression profiles led to the identification of a regulatory network which includes the miRNAs up-regulated by FHC silencing, as well as91 down-regulated putative target genes. These genes were further classified in 9 networks; the highest scoring network, "Cell Death and Survival, Hematological System Development and Function, Hematopoiesis", is composed by 18 focus molecules including RAF1 and ERK1/2. We confirmed that, following FHC silencing, ERK1/2 phosphorylation is severely impaired and that RAF1 mRNA is significantly down-regulated. Taken all together, our data indicate that, in our experimental model, FHC silencing may affect RAF1/pERK1/2 levels through the modulation of a specific set of miRNAs and add new insights in to the relationship among iron homeostasis and miRNAs.
Conflict of interest statement
Figures



References
-
- Beard JL, Connor JR, Jones BC. (1993) Iron in the brain.Nutr Rev 51: 157–170. - PubMed
-
- Levi S, Luzzago A, Cesareni G, Cozzi A, Franceschinelli F. (1988) Mechanismofferritinironuptake:activityof theH-chainanddeletionmappingof theferro-oxidasesite.Astudy ofironuptakeandferro-oxidaseactivityofhumanliver,recombinantH-chainferritins, and oftwoH-chain deletionmutants. J Biol Chem 263(34):18086–18092. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

