Methylxanthines Increase Expression of the Splicing Factor SRSF2 by Regulating Multiple Post-transcriptional Mechanisms
- PMID: 25818199
- PMCID: PMC4463444
- DOI: 10.1074/jbc.M114.624254
Methylxanthines Increase Expression of the Splicing Factor SRSF2 by Regulating Multiple Post-transcriptional Mechanisms
Abstract
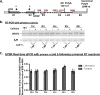
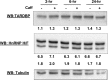
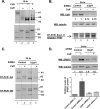
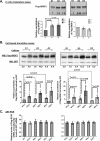
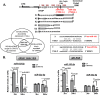
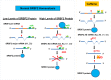
We have previously reported that the methylxanthine caffeine increases expression of the splicing factor SRSF2, the levels of which are normally controlled by a negative autoregulatory loop. In the present study we have investigated the mechanisms by which methylxanthines induce this aberrant overexpression. RT-PCR analyses suggested little impact of caffeine on SRSF2 total mRNA levels. Instead, caffeine induced changes in the levels of SRSF2 3' UTR splice variants. Although some of these variants were substrates for nonsense-medicated decay (NMD), and could potentially have been stabilized by caffeine-mediated inhibition of NMD, down-regulation of NMD by a genetic approach was not sufficient to reproduce the phenotype. Furthermore, cell-based assays demonstrated that some of the caffeine-induced variants were intrinsically more efficiently translated than others; the addition of caffeine increased the translational efficiency of most SRSF2 transcripts. MicroRNA array analyses revealed a significant caffeine-mediated decrease in the expression of two SRSF2-targeting miRs, both of which were shown to repress translation of specific SRSF2 splice variants. These data support a complex model whereby caffeine down-regulates SRSF2-targeting microRNAs, leading to an increase in SRSF2 translation, which in turn induces SRSF2 splicing. SRSF2 splice variants are then stabilized by caffeine-mediated NMD inhibition, breaking the normal negative feedback loop and allowing the aberrant increase in SRSF2 protein levels. These findings highlight the complexity of SRSF2 gene regulation, and suggest ways in which SRSF2 expression may be dysregulated in disease.
Keywords: alternative splicing; caffeine, SRSF2, nonsense-mediated decay; gene regulation; homeostasis; microRNA (miRNA); translation regulation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Similar articles
-
Splicing factor SRSF2-centric gene regulation.Int J Biol Sci. 2021 Apr 16;17(7):1708-1715. doi: 10.7150/ijbs.58888. eCollection 2021. Int J Biol Sci. 2021. PMID: 33994855 Free PMC article. Review.
-
Caffeine regulates alternative splicing in a subset of cancer-associated genes: a role for SC35.Mol Cell Biol. 2008 Jan;28(2):883-95. doi: 10.1128/MCB.01345-07. Epub 2007 Nov 19. Mol Cell Biol. 2008. PMID: 18025108 Free PMC article.
-
Recurrent SRSF2 mutations in MDS affect both splicing and NMD.Genes Dev. 2020 Mar 1;34(5-6):413-427. doi: 10.1101/gad.332270.119. Epub 2020 Jan 30. Genes Dev. 2020. PMID: 32001512 Free PMC article.
-
Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities.Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):E4726-34. doi: 10.1073/pnas.1514105112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261309 Free PMC article.
-
Alternative splicing and nonsense-mediated mRNA decay enforce neural specific gene expression.Int J Dev Neurosci. 2016 Dec;55:102-108. doi: 10.1016/j.ijdevneu.2016.03.003. Epub 2016 Mar 8. Int J Dev Neurosci. 2016. PMID: 26968265 Free PMC article. Review.
Cited by
-
The Epigenetic Effects of Coffee.Molecules. 2023 Feb 13;28(4):1770. doi: 10.3390/molecules28041770. Molecules. 2023. PMID: 36838754 Free PMC article. Review.
-
Serum miRNAs are differentially altered by ethanol and caffeine consumption in rats.Toxicol Res (Camb). 2019 Jul 17;8(6):842-849. doi: 10.1039/c9tx00069k. eCollection 2019 Nov 1. Toxicol Res (Camb). 2019. PMID: 32055392 Free PMC article.
-
Splicing to keep splicing: A feedback system for cellular homeostasis and state transition.Clin Transl Med. 2025 Jun;15(6):e70369. doi: 10.1002/ctm2.70369. Clin Transl Med. 2025. PMID: 40538061 Free PMC article. Review.
-
Towards Splicing Therapy for Lysosomal Storage Disorders: Methylxanthines and Luteolin Ameliorate Splicing Defects in Aspartylglucosaminuria and Classic Late Infantile Neuronal Ceroid Lipofuscinosis.Cells. 2021 Oct 20;10(11):2813. doi: 10.3390/cells10112813. Cells. 2021. PMID: 34831035 Free PMC article.
-
Splicing factor SRSF2-centric gene regulation.Int J Biol Sci. 2021 Apr 16;17(7):1708-1715. doi: 10.7150/ijbs.58888. eCollection 2021. Int J Biol Sci. 2021. PMID: 33994855 Free PMC article. Review.
References
-
- McGlincy N. J., Smith C. W. (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 33, 385–393 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

