Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing
- PMID: 25818644
- PMCID: PMC4402144
- DOI: 10.1016/j.molcel.2015.02.023
Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing
Abstract
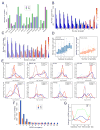
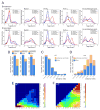
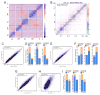
Chromosomes of metazoan organisms are partitioned in the interphase nucleus into discrete topologically associating domains (TADs). Borders between TADs are formed in regions containing active genes and clusters of architectural protein binding sites. The transcription of most genes is repressed after temperature stress in Drosophila. Here we show that temperature stress induces relocalization of architectural proteins from TAD borders to inside TADs, and this is accompanied by a dramatic rearrangement in the 3D organization of the nucleus. TAD border strength declines, allowing for an increase in long-distance inter-TAD interactions. Similar but quantitatively weaker effects are observed upon inhibition of transcription or depletion of individual architectural proteins. Heat shock-induced inter-TAD interactions result in increased contacts among enhancers and promoters of silenced genes, which recruit Pc and form Pc bodies in the nucleolus. These results suggest that the TAD organization of metazoan genomes is plastic and can be reconfigured quickly.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




References
-
- Cheutin T, Cavalli G. Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Current opinion in genetics & development. 2014;25:30–37. - PubMed
-
- Delest A, Sexton T, Cavalli G. Polycomb: a paradigm for genome organization from one to three dimensions. Current opinion in cell biology. 2012;24:405–414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

