Collateral sensitivity of antibiotic-resistant microbes
- PMID: 25818802
- PMCID: PMC5958998
- DOI: 10.1016/j.tim.2015.02.009
Collateral sensitivity of antibiotic-resistant microbes
Abstract
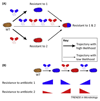
Understanding how evolution of microbial resistance towards a given antibiotic influences susceptibility to other drugs is a challenge of profound importance. By combining laboratory evolution, genome sequencing, and functional analyses, recent works have charted the map of evolutionary trade-offs between antibiotics and have explored the underlying molecular mechanisms. Strikingly, mutations that caused multidrug resistance in bacteria simultaneously enhanced sensitivity to many other unrelated drugs (collateral sensitivity). Here, we explore how this emerging research sheds new light on resistance mechanisms and the way it could be exploited for the development of alternative antimicrobial strategies.
Keywords: antibiotic resistance; collateral sensitivity; cross-resistance; experimental evolution; multidrug resistance.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


References
-
- Velicer GJ, et al. Application of traditional and phylogenetically based comparative methods to test for a trade-off in bacterial growth rate at low versus high substrate concentration. Microb Ecol. 1999;38:191–200. - PubMed
-
- Gagneux S, et al. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–1946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

