An Rtf2 Domain-Containing Protein Influences Pre-mRNA Splicing and Is Essential for Embryonic Development in Arabidopsis thaliana
- PMID: 25819795
- PMCID: PMC4492377
- DOI: 10.1534/genetics.115.176438
An Rtf2 Domain-Containing Protein Influences Pre-mRNA Splicing and Is Essential for Embryonic Development in Arabidopsis thaliana
Abstract
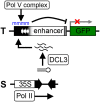
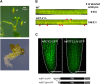
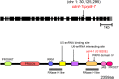
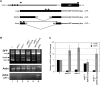
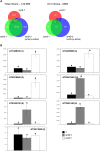
Alternative splicing is prevalent in plants, but little is known about its regulation in the context of developmental and signaling pathways. We describe here a new factor that influences pre-messengerRNA (mRNA) splicing and is essential for embryonic development in Arabidopsis thaliana. This factor was retrieved in a genetic screen that identified mutants impaired in expression of an alternatively spliced GFP reporter gene. In addition to the known spliceosomal component PRP8, the screen recovered Arabidopsis RTF2 (AtRTF2), a previously uncharacterized, evolutionarily conserved protein containing a replication termination factor 2 (Rtf2) domain. A homozygous null mutation in AtRTF2 is embryo lethal, indicating that AtRTF2 is an essential protein. Quantitative RT-PCR demonstrated that impaired expression of GFP in atrtf2 and prp8 mutants is due to inefficient splicing of the GFP pre-mRNA. A genome-wide analysis using RNA sequencing indicated that 13-16% of total introns are retained to a significant degree in atrtf2 mutants. Considering these results and previous suggestions that Rtf2 represents an ubiquitin-related domain, we discuss the possible role of AtRTF2 in ubiquitin-based regulation of pre-mRNA splicing.
Keywords: C2HC2 zinc finger; Rtf2 domain; alternative splicing; intron retention; ubiquitin ligase.
Copyright © 2015 by the Genetics Society of America.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

