Bridging the gap between postembryonic cell lineages and identified embryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster
- PMID: 25819843
- PMCID: PMC4400586
- DOI: 10.1242/bio.201411072
Bridging the gap between postembryonic cell lineages and identified embryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster
Abstract
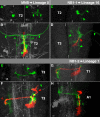
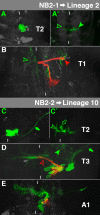
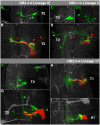
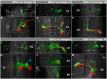
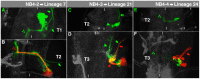
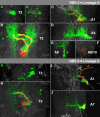
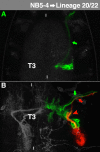
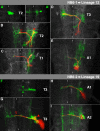
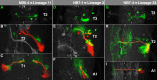
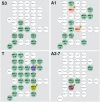
The clarification of complete cell lineages, which are produced by specific stem cells, is fundamental for understanding mechanisms, controlling the generation of cell diversity and patterning in an emerging tissue. In the developing Central Nervous System (CNS) of Drosophila, neural stem cells (neuroblasts) exhibit two periods of proliferation: During embryogenesis they produce primary lineages, which form the larval CNS. After a phase of mitotic quiescence, a subpopulation of them resumes proliferation in the larva to give rise to secondary lineages that build up the CNS of the adult fly. Within the ventral nerve cord (VNC) detailed descriptions exist for both primary and secondary lineages. However, while primary lineages have been linked to identified neuroblasts, the assignment of secondary lineages has so far been hampered by technical limitations. Therefore, primary and secondary neural lineages co-existed as isolated model systems. Here we provide the missing link between the two systems for all lineages in the thoracic and abdominal neuromeres. Using the Flybow technique, embryonic neuroblasts were identified by their characteristic and unique lineages in the living embryo and their further development was traced into the late larval stage. This comprehensive analysis provides the first complete view of which embryonic neuroblasts are postembryonically reactivated along the anterior/posterior-axis of the VNC, and reveals the relationship between projection patterns of primary and secondary sublineages.
Keywords: CNS development; Cell lineage; Drosophila; Flybow; Neuroblast; Segmental patterning.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

