Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in the obligate root parasitic plant, Phelipanche ramosa L. Pomel
- PMID: 25821070
- PMCID: PMC4449535
- DOI: 10.1093/jxb/erv119
Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in the obligate root parasitic plant, Phelipanche ramosa L. Pomel
Abstract
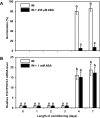
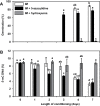
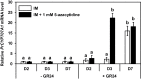
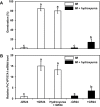
Seed dormancy release of the obligate root parasitic plant, Phelipanche ramosa, requires a minimum 4-day conditioning period followed by stimulation by host-derived germination stimulants, such as strigolactones. Germination is then mediated by germination stimulant-dependent activation of PrCYP707A1, an abscisic acid catabolic gene. The molecular mechanisms occurring during the conditioning period that silence PrCYP707A1 expression and regulate germination stimulant response are almost unknown. Here, global DNA methylation quantification associated with pharmacological approaches and cytosine methylation analysis of the PrCYP707A1 promoter were used to investigate the modulation and possible role of DNA methylation during the conditioning period and in the PrCYP707A1 response to GR24, a synthetic strigolactone analogue. Active global DNA demethylation occurs during the conditioning period and is required for PrCYP707A1 activation by GR24 and for subsequent seed germination. Treatment with 5-azacytidine, a DNA-hypomethylating molecule, reduces the length of the conditioning period. Conversely, hydroxyurea, a hypermethylating agent, inhibits PrCYP707A1 expression and seed germination. Methylated DNA immunoprecipitation followed by PCR experiments and bisulfite sequencing revealed that DNA demethylation particularly impacts a 78-nucleotide sequence in the PrCYP707A1 promoter. The results here demonstrate that the DNA methylation status during the conditioning period plays a crucial role independently of abscisic acid in the regulation of P. ramosa seed germination by controlling the strigolactone-dependent expression of PrCYP707A1.
Keywords: Bisulfite sequencing; DNA methylation; branched broomrape (Phelipanche ramosa); dormancy release; epigenetic regulation; parasitic plant; seed germination; strigolactone..
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures
Similar articles
-
PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24.J Exp Bot. 2012 Sep;63(14):5311-22. doi: 10.1093/jxb/ers189. Epub 2012 Aug 1. J Exp Bot. 2012. PMID: 22859674 Free PMC article.
-
An improved axenic system for studying pre-infection development of the parasitic plant Orobanche ramosa.Ann Bot. 2005 Nov;96(6):1121-7. doi: 10.1093/aob/mci263. Epub 2005 Sep 12. Ann Bot. 2005. PMID: 16157629 Free PMC article.
-
Strigolactone deficiency confers resistance in tomato line SL-ORT1 to the parasitic weeds Phelipanche and Orobanche spp.Phytopathology. 2011 Feb;101(2):213-22. doi: 10.1094/PHYTO-07-10-0184. Phytopathology. 2011. PMID: 20942651
-
Structure and function of natural and synthetic signalling molecules in parasitic weed germination.Pest Manag Sci. 2009 May;65(5):478-91. doi: 10.1002/ps.1706. Pest Manag Sci. 2009. PMID: 19222046 Review.
-
Noncanonical Strigolactone Analogues Highlight Selectivity for Stimulating Germination in Two Phelipanche ramosa Populations.J Nat Prod. 2022 Aug 26;85(8):1976-1992. doi: 10.1021/acs.jnatprod.2c00282. Epub 2022 Jul 1. J Nat Prod. 2022. PMID: 35776904 Review.
Cited by
-
Emerging Roles of Strigolactones in Plant Responses to Stress and Development.Front Plant Sci. 2016 Apr 5;7:434. doi: 10.3389/fpls.2016.00434. eCollection 2016. Front Plant Sci. 2016. PMID: 27092155 Free PMC article. Review.
-
The effect of nojirimycin on the transcriptome of germinating Orobanche minor seeds.J Pestic Sci. 2020 Nov 20;45(4):230-237. doi: 10.1584/jpestics.D20-057. J Pestic Sci. 2020. PMID: 33304192 Free PMC article.
-
Karrikin signalling: impacts on plant development and abiotic stress tolerance.J Exp Bot. 2024 Feb 12;75(4):1174-1186. doi: 10.1093/jxb/erad476. J Exp Bot. 2024. PMID: 38001035 Free PMC article.
-
Genomic and Epigenomic Mechanisms of the Interaction between Parasitic and Host Plants.Int J Mol Sci. 2023 Jan 31;24(3):2647. doi: 10.3390/ijms24032647. Int J Mol Sci. 2023. PMID: 36768970 Free PMC article. Review.
-
Effect of Arsenic Soil Contamination on Stress Response Metabolites, 5-Methylcytosine Level and CDC25 Expression in Spinach.Toxics. 2023 Jun 29;11(7):568. doi: 10.3390/toxics11070568. Toxics. 2023. PMID: 37505533 Free PMC article.
References
-
- Bar-Nun N, Mayer AM. 1993. Preconditioning and germination of Orobanche seeds: respiration and protein synthesis. Phytochemistry 34, 39–45.
-
- Bar-Nun N, Mayer AM. 2002. Composition of and changes in storage compounds in Orobanche aegyptiaca seeds during preconditioning. Israel Journal of Plant Sciences 50, 278–279.
-
- Bar-Nun N, Plakhine D, Joel DM, Mayer AM. 2003. Changes in the activity of the alternative oxidase in Orobanche seeds during conditioning and their possible physiological function. Phytochemistry 64, 235–241. - PubMed
-
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes & Development 16, 6–21. - PubMed
-
- Boyes J, Bird A. 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64, 1123–1134. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

