Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma
- PMID: 25821520
- PMCID: PMC4375928
- DOI: 10.1186/s13073-015-0142-6
Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma
Abstract
Background: Long non-coding RNAs (lncRNAs) are emerging as molecules that significantly impact many cellular processes and have been associated with almost every human cancer. Compared to protein-coding genes, lncRNA genes are often associated with transposable elements, particularly with endogenous retroviral elements (ERVs). ERVs can have potentially deleterious effects on genome structure and function, so these elements are typically silenced in normal somatic tissues, albeit with varying efficiency. The aberrant regulation of ERVs associated with lncRNAs (ERV-lncRNAs), coupled with the diverse range of lncRNA functions, creates significant potential for ERV-lncRNAs to impact cancer biology.
Methods: We used RNA-seq analysis to identify and profile the expression of a novel lncRNA in six large cohorts, including over 7,500 samples from The Cancer Genome Atlas (TCGA).
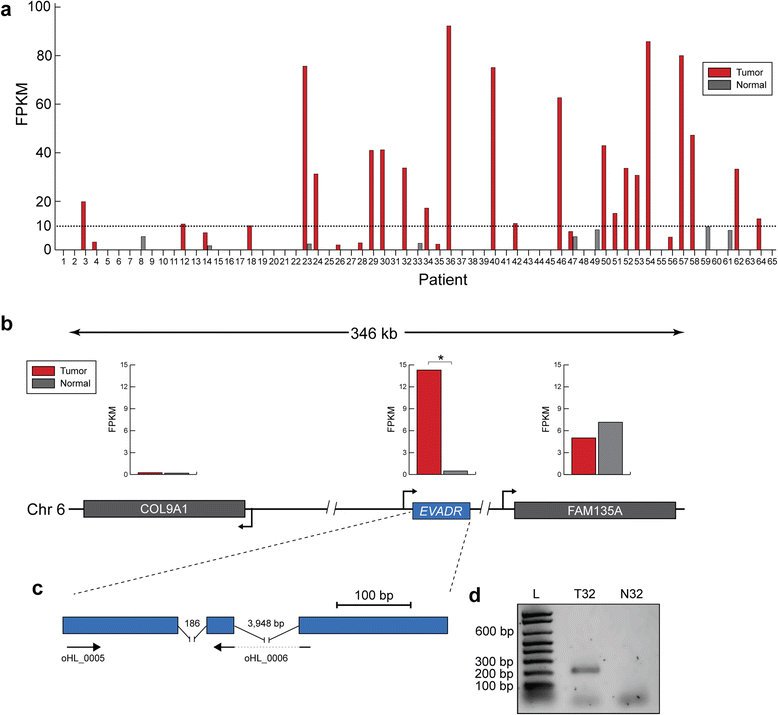
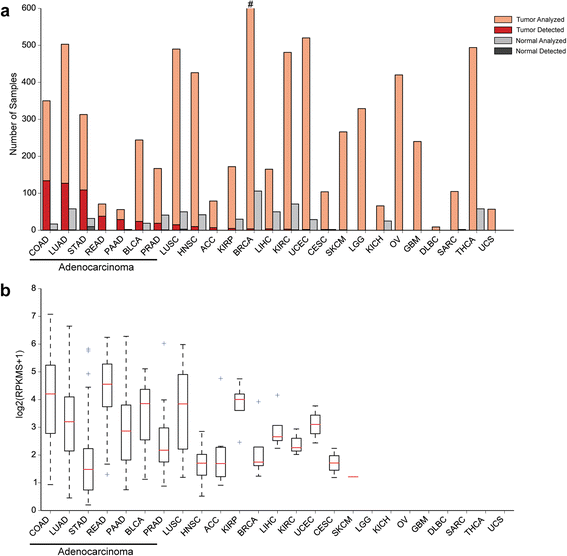
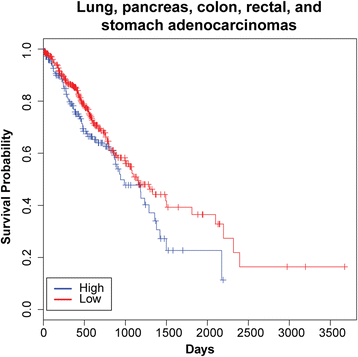
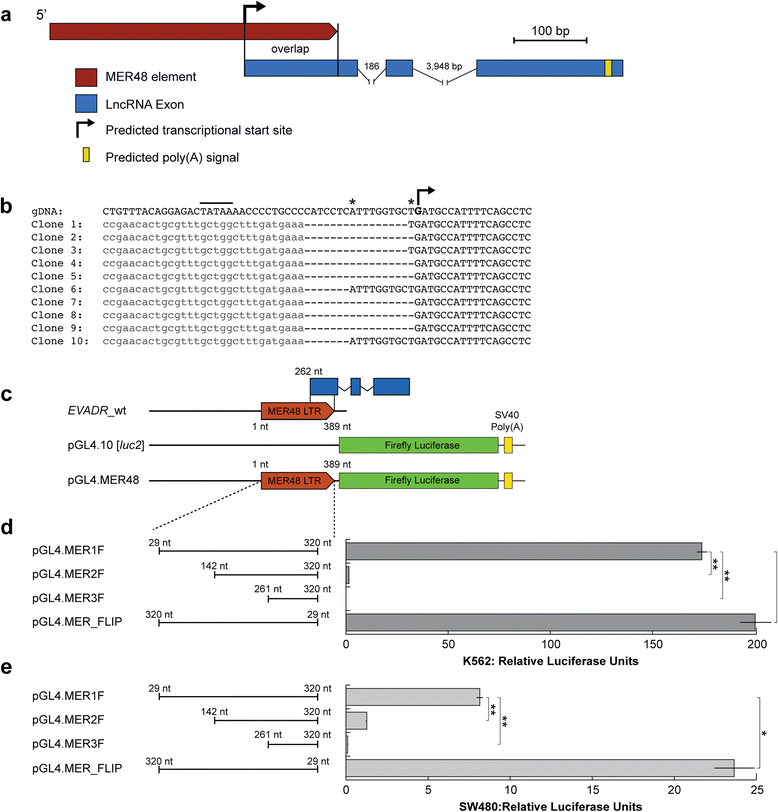
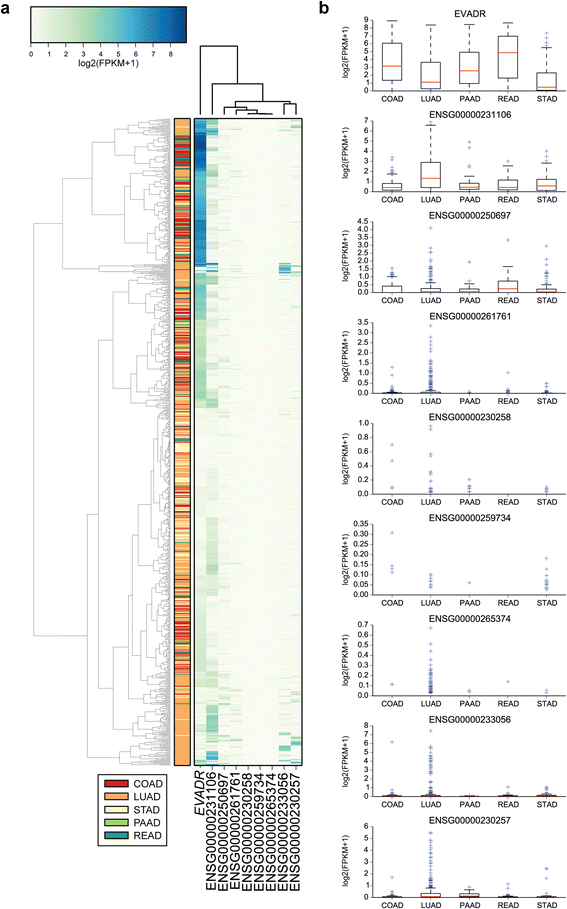
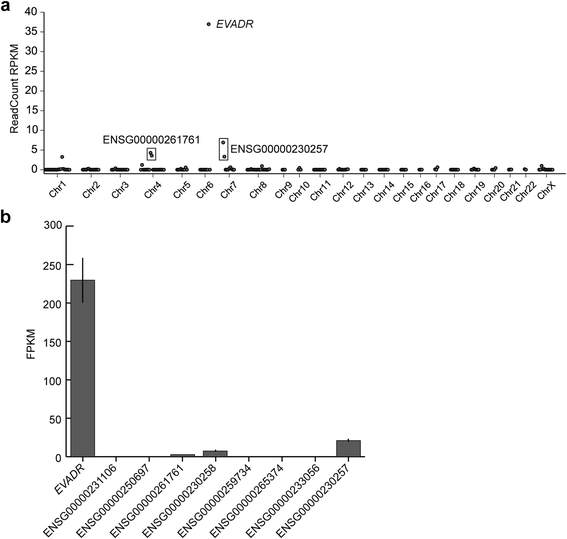
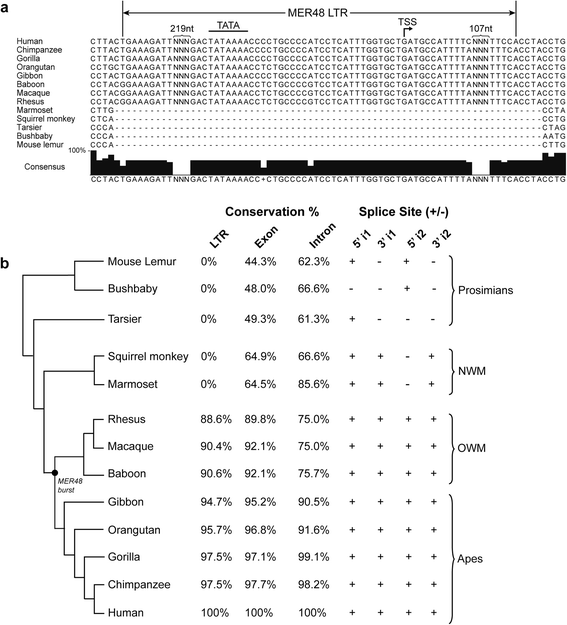
Results: We identified the tumor-specific expression of a novel lncRNA that we have named Endogenous retroViral-associated ADenocarcinoma RNA or 'EVADR', by analyzing RNA-seq data derived from colorectal tumors and matched normal control tissues. Subsequent analysis of TCGA RNA-seq data revealed the striking association of EVADR with adenocarcinomas, which are tumors of glandular origin. Moderate to high levels of EVADR were detected in 25 to 53% of colon, rectal, lung, pancreas and stomach adenocarcinomas (mean = 30 to 144 FPKM), and EVADR expression correlated with decreased patient survival (Cox regression; hazard ratio = 1.47, 95% confidence interval = 1.06 to 2.04, P = 0.02). In tumor sites of non-glandular origin, EVADR expression was detectable at only very low levels and in less than 10% of patients. For EVADR, a MER48 ERV element provides an active promoter to drive its transcription. Genome-wide, MER48 insertions are associated with nine lncRNAs, but none of the MER48-associated lncRNAs other than EVADR were consistently expressed in adenocarcinomas, demonstrating the specific activation of EVADR. The sequence and structure of the EVADR locus is highly conserved among Old World monkeys and apes but not New World monkeys or prosimians, where the MER48 insertion is absent. Conservation of the EVADR locus suggests a functional role for this novel lncRNA in humans and our closest primate relatives.
Conclusions: Our results describe the specific activation of a highly conserved ERV-lncRNA in numerous cancers of glandular origin, a finding with diagnostic, prognostic and therapeutic implications.
Figures
Similar articles
-
Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma.Genome Med. 2015 Nov 1;7:110. doi: 10.1186/s13073-015-0230-7. Genome Med. 2015. PMID: 26521025 Free PMC article.
-
Endogenous Retrovirus-Derived Long Noncoding RNA Enhances Innate Immune Responses via Derepressing RELA Expression.mBio. 2019 Jul 30;10(4):e00937-19. doi: 10.1128/mBio.00937-19. mBio. 2019. PMID: 31363026 Free PMC article.
-
Three-lncRNA signature is a potential prognostic biomarker for pancreatic adenocarcinoma.Oncotarget. 2018 Feb 8;9(36):24248-24259. doi: 10.18632/oncotarget.24443. eCollection 2018 May 11. Oncotarget. 2018. PMID: 29849937 Free PMC article.
-
Targeting long non-coding RNAs in cancers: progress and prospects.Int J Biochem Cell Biol. 2013 Aug;45(8):1895-910. doi: 10.1016/j.biocel.2013.05.030. Epub 2013 Jun 4. Int J Biochem Cell Biol. 2013. PMID: 23748105 Review.
-
Expression Specificity of Disease-Associated lncRNAs: Toward Personalized Medicine.Curr Top Microbiol Immunol. 2016;394:237-58. doi: 10.1007/82_2015_464. Curr Top Microbiol Immunol. 2016. PMID: 26318140 Review.
Cited by
-
Long non-coding RNA ASncmtRNA-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells.Exp Ther Med. 2017 Feb;13(2):581-587. doi: 10.3892/etm.2017.4027. Epub 2017 Jan 5. Exp Ther Med. 2017. PMID: 28352334 Free PMC article.
-
The Expression of lncRNAs EVADR and LUESCC in Colorectal Tumor Tissues and Their Association With the CRC Risk.Cancer Rep (Hoboken). 2025 Jul;8(7):e70232. doi: 10.1002/cnr2.70232. Cancer Rep (Hoboken). 2025. PMID: 40620190 Free PMC article.
-
Locus-Specific Characterization of Human Endogenous Retrovirus Expression in Prostate, Breast, and Colon Cancers.Cancer Res. 2021 Jul 1;81(13):3449-3460. doi: 10.1158/0008-5472.CAN-20-3975. Epub 2021 May 3. Cancer Res. 2021. PMID: 33941616 Free PMC article.
-
KDM3A Ablation Activates Endogenous Retrovirus Expression to Stimulate Antitumor Immunity in Gastric Cancer.Adv Sci (Weinh). 2024 Oct;11(40):e2309983. doi: 10.1002/advs.202309983. Epub 2024 Jul 19. Adv Sci (Weinh). 2024. PMID: 39031630 Free PMC article.
-
Human Endogenous Retrovirus-H-Derived miR-4454 Inhibits the Expression of DNAJB4 and SASH1 in Non-Muscle-Invasive Bladder Cancer.Genes (Basel). 2023 Jul 7;14(7):1410. doi: 10.3390/genes14071410. Genes (Basel). 2023. PMID: 37510314 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

