Identification of real microRNA precursors with a pseudo structure status composition approach
- PMID: 25821974
- PMCID: PMC4378912
- DOI: 10.1371/journal.pone.0121501
Identification of real microRNA precursors with a pseudo structure status composition approach
Abstract
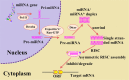
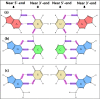
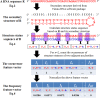
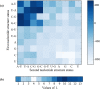
Containing about 22 nucleotides, a micro RNA (abbreviated miRNA) is a small non-coding RNA molecule, functioning in transcriptional and post-transcriptional regulation of gene expression. The human genome may encode over 1000 miRNAs. Albeit poorly characterized, miRNAs are widely deemed as important regulators of biological processes. Aberrant expression of miRNAs has been observed in many cancers and other disease states, indicating they are deeply implicated with these diseases, particularly in carcinogenesis. Therefore, it is important for both basic research and miRNA-based therapy to discriminate the real pre-miRNAs from the false ones (such as hairpin sequences with similar stem-loops). Particularly, with the avalanche of RNA sequences generated in the postgenomic age, it is highly desired to develop computational sequence-based methods in this regard. Here two new predictors, called "iMcRNA-PseSSC" and "iMcRNA-ExPseSSC", were proposed for identifying the human pre-microRNAs by incorporating the global or long-range structure-order information using a way quite similar to the pseudo amino acid composition approach. Rigorous cross-validations on a much larger and more stringent newly constructed benchmark dataset showed that the two new predictors (accessible at http://bioinformatics.hitsz.edu.cn/iMcRNA/) outperformed or were highly comparable with the best existing predictors in this area.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

