Testing for genetic associations in arbitrarily structured populations
- PMID: 25822090
- PMCID: PMC4464830
- DOI: 10.1038/ng.3244
Testing for genetic associations in arbitrarily structured populations
Abstract
We present a new statistical test of association between a trait and genetic markers, which we theoretically and practically prove to be robust to arbitrarily complex population structure. The statistical test involves a set of parameters that can be directly estimated from large-scale genotyping data, such as those measured in genome-wide association studies (GWAS). We also derive a new set of methodologies, called a 'genotype-conditional association test' (GCAT), shown to provide accurate association tests in populations with complex structures, manifested in both the genetic and non-genetic contributions to the trait. We demonstrate the proposed method on a large simulation study and on the Northern Finland Birth Cohort study. In the Finland study, we identify several new significant loci that other methods do not detect. Our proposed framework provides a substantially different approach to the problem from existing methods, such as the linear mixed-model and principal-component approaches.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
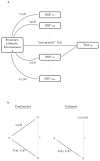
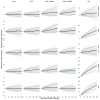
Similar articles
-
Principal component regression and linear mixed model in association analysis of structured samples: competitors or complements?Genet Epidemiol. 2015 Mar;39(3):149-55. doi: 10.1002/gepi.21879. Epub 2014 Dec 23. Genet Epidemiol. 2015. PMID: 25536929 Free PMC article.
-
A mixed-model approach for genome-wide association studies of correlated traits in structured populations.Nat Genet. 2012 Sep;44(9):1066-71. doi: 10.1038/ng.2376. Epub 2012 Aug 19. Nat Genet. 2012. PMID: 22902788 Free PMC article.
-
Multiple phenotype association tests using summary statistics in genome-wide association studies.Biometrics. 2018 Mar;74(1):165-175. doi: 10.1111/biom.12735. Epub 2017 Jun 26. Biometrics. 2018. PMID: 28653391 Free PMC article.
-
Variance component model to account for sample structure in genome-wide association studies.Nat Genet. 2010 Apr;42(4):348-54. doi: 10.1038/ng.548. Epub 2010 Mar 7. Nat Genet. 2010. PMID: 20208533 Free PMC article.
-
Population structure in genetic studies: Confounding factors and mixed models.PLoS Genet. 2018 Dec 27;14(12):e1007309. doi: 10.1371/journal.pgen.1007309. eCollection 2018 Dec. PLoS Genet. 2018. PMID: 30589851 Free PMC article. Review.
Cited by
-
Identifying latent genetic interactions in genome-wide association studies using multiple traits.Genome Med. 2024 Apr 25;16(1):62. doi: 10.1186/s13073-024-01329-0. Genome Med. 2024. PMID: 38664839 Free PMC article.
-
A Likelihood-Free Estimator of Population Structure Bridging Admixture Models and Principal Components Analysis.Genetics. 2019 Aug;212(4):1009-1029. doi: 10.1534/genetics.119.302159. Epub 2019 Apr 26. Genetics. 2019. PMID: 31028112 Free PMC article.
-
Regenie.QRS: computationally efficient whole-genome quantile regression at biobank scale.bioRxiv [Preprint]. 2025 May 7:2025.05.02.651730. doi: 10.1101/2025.05.02.651730. bioRxiv. 2025. PMID: 40654860 Free PMC article. Preprint.
-
Gene-Based Association Testing of Dichotomous Traits With Generalized Functional Linear Mixed Models Using Extended Pedigrees: Applications to Age-Related Macular Degeneration.J Am Stat Assoc. 2021;116(534):531-545. doi: 10.1080/01621459.2020.1799809. Epub 2020 Jul 28. J Am Stat Assoc. 2021. PMID: 34321704 Free PMC article.
-
A robust two-sample transcriptome-wide Mendelian randomization method integrating GWAS with multi-tissue eQTL summary statistics.Genet Epidemiol. 2021 Jun;45(4):353-371. doi: 10.1002/gepi.22380. Epub 2021 Apr 9. Genet Epidemiol. 2021. PMID: 33834509 Free PMC article.
References
-
- McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. - PubMed
-
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–251. - PubMed
-
- Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies. Stat Sci. 2009;24:451–471.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

