DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia
- PMID: 25822366
- PMCID: PMC4390532
- DOI: 10.1038/nm.3832
DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia
Abstract
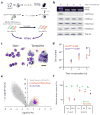
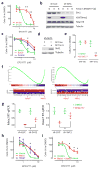
Rearrangements of MLL (encoding lysine-specific methyltransferase 2A and officially known as KMT2A; herein referred to as MLL to denote the gene associated with mixed-lineage leukemia) generate MLL fusion proteins that bind DNA and drive leukemogenic gene expression. This gene expression program is dependent on the disruptor of telomeric silencing 1-like histone 3 lysine 79 (H3K79) methyltransferase DOT1L, and small-molecule DOT1L inhibitors show promise as therapeutics for these leukemias. However, the mechanisms underlying this dependency are unclear. We conducted a genome-scale RNAi screen and found that the histone deacetylase SIRT1 is required for the establishment of a heterochromatin-like state around MLL fusion target genes after DOT1L inhibition. DOT1L inhibits chromatin localization of a repressive complex composed of SIRT1 and the H3K9 methyltransferase SUV39H1, thereby maintaining an open chromatin state with elevated H3K9 acetylation and minimal H3K9 methylation at MLL fusion target genes. Furthermore, the combination of SIRT1 activators and DOT1L inhibitors shows enhanced antiproliferative activity against MLL-rearranged leukemia cells. These results indicate that the dynamic interplay between chromatin regulators controlling the activation and repression of gene expression could provide novel opportunities for combination therapy.
Conflict of interest statement
S.A.A. is a consultant for Epizyme Inc.. The remaining authors report no competing financial interests.
Figures




Comment in
-
Harnessing gene repression to inhibit leukemia.Nat Med. 2015 Apr;21(4):308-10. doi: 10.1038/nm.3834. Nat Med. 2015. PMID: 25849269 No abstract available.
References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nature reviews Molecular cell biology. 2014;15:703–708. - PubMed
-
- Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. - PubMed
-
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

