Properties of protein drug target classes
- PMID: 25822509
- PMCID: PMC4379170
- DOI: 10.1371/journal.pone.0117955
Properties of protein drug target classes
Abstract
Accurate identification of drug targets is a crucial part of any drug development program. We mined the human proteome to discover properties of proteins that may be important in determining their suitability for pharmaceutical modulation. Data was gathered concerning each protein's sequence, post-translational modifications, secondary structure, germline variants, expression profile and drug target status. The data was then analysed to determine features for which the target and non-target proteins had significantly different values. This analysis was repeated for subsets of the proteome consisting of all G-protein coupled receptors, ion channels, kinases and proteases, as well as proteins that are implicated in cancer. Machine learning was used to quantify the proteins in each dataset in terms of their potential to serve as a drug target. This was accomplished by first inducing a random forest that could distinguish between its targets and non-targets, and then using the random forest to quantify the drug target likeness of the non-targets. The properties that can best differentiate targets from non-targets were primarily those that are directly related to a protein's sequence (e.g. secondary structure). Germline variants, expression levels and interactions between proteins had minimal discriminative power. Overall, the best indicators of drug target likeness were found to be the proteins' hydrophobicities, in vivo half-lives, propensity for being membrane bound and the fraction of non-polar amino acids in their sequences. In terms of predicting potential targets, datasets of proteases, ion channels and cancer proteins were able to induce random forests that were highly capable of distinguishing between targets and non-targets. The non-target proteins predicted to be targets by these random forests comprise the set of the most suitable potential future drug targets, and should therefore be prioritised when building a drug development programme.
Conflict of interest statement
Figures
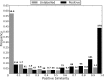

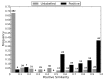
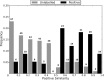
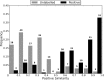
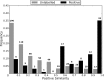
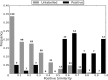
References
-
- Hopkins AL, Groom CR (2002) The druggable genome. Nature Rev Drug Discovery 1: 727–730. - PubMed
-
- Xu H, Xu HY, Lin MZ, Wang W, Li ZM, et al. (2007) Learning the drug target-likeness of a protein. Proteomics 7: 4255–4263. - PubMed
-
- Chen XP, Du GH (2007) Target validation: A door to drug discovery. Drug discoveries & therapeutics 1: 23–29. - PubMed
-
- Drews J (2000) Drug discovery: A historical perspective. Science 287: 1960–1964. - PubMed
-
- Imming P, Sinning S, Meyer A (2006) Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov 5: 821–834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

