Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis
- PMID: 25824154
- PMCID: PMC4389249
- DOI: 10.1038/ncomms7740
Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis
Erratum in
-
Erratum: Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis.Nat Commun. 2015 May 12;6:7272. doi: 10.1038/ncomms8272. Nat Commun. 2015. PMID: 25966020 Free PMC article. No abstract available.
-
Publisher Correction: Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis.Nat Commun. 2019 Nov 22;10(1):5326. doi: 10.1038/s41467-019-13138-w. Nat Commun. 2019. PMID: 31757940 Free PMC article.
Abstract
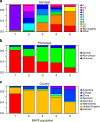
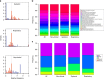
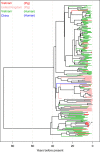
Streptococcus suis causes disease in pigs worldwide and is increasingly implicated in zoonotic disease in East and South-East Asia. To understand the genetic basis of disease in S. suis, we study the genomes of 375 isolates with detailed clinical phenotypes from pigs and humans from the United Kingdom and Vietnam. Here, we show that isolates associated with disease contain substantially fewer genes than non-clinical isolates, but are more likely to encode virulence factors. Human disease isolates are limited to a single-virulent population, originating in the 1920, s when pig production was intensified, but no consistent genomic differences between pig and human isolates are observed. There is little geographical clustering of different S. suis subpopulations, and the bacterium undergoes high rates of recombination, implying that an increase in virulence anywhere in the world could have a global impact over a short timescale.
Figures

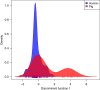
References
-
- Wertheim H. F. L., Nghia H. D. T., Taylor W. & Schultsz C. Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48, 617–625 (2009) . - PubMed
-
- Vilaichone R.-K., Vilaichone W., Nunthapisud P. & Wilde H. Streptococcus suis infection in Thailand. J. Med. Assoc. Thailand 85, S109–S117 (2002) . - PubMed
Publication types
MeSH terms
Substances
Associated data
- SRA/ERS018463
- SRA/ERS132352
- SRA/ERS132353
- SRA/ERS132354
- SRA/ERS132355
- SRA/ERS132356
- SRA/ERS132357
- SRA/ERS132358
- SRA/ERS132359
- SRA/ERS132360
- SRA/ERS132361
- SRA/ERS132362
- SRA/ERS132363
- SRA/ERS132364
- SRA/ERS132365
- SRA/ERS132366
- SRA/ERS132367
- SRA/ERS132370
- SRA/ERS132371
- SRA/ERS132372
- SRA/ERS132373
- SRA/ERS132374
- SRA/ERS132375
- SRA/ERS132376
- SRA/ERS132380
- SRA/ERS132382
- SRA/ERS132383
- SRA/ERS132384
- SRA/ERS132385
- SRA/ERS132386
- SRA/ERS132387
- SRA/ERS132388
- SRA/ERS132389
- SRA/ERS132390
- SRA/ERS132391
- SRA/ERS132392
- SRA/ERS132393
- SRA/ERS132394
- SRA/ERS132395
- SRA/ERS132396
- SRA/ERS132397
- SRA/ERS132398
- SRA/ERS132399
- SRA/ERS132400
- SRA/ERS132401
- SRA/ERS132402
- SRA/ERS132403
- SRA/ERS132404
- SRA/ERS132406
- SRA/ERS132409
- SRA/ERS132410
- SRA/ERS132411
- SRA/ERS132412
- SRA/ERS132413
- SRA/ERS132414
- SRA/ERS132415
- SRA/ERS132416
- SRA/ERS132417
- SRA/ERS132418
- SRA/ERS132419
- SRA/ERS132420
- SRA/ERS132421
- SRA/ERS132422
- SRA/ERS132423
- SRA/ERS132424
- SRA/ERS132425
- SRA/ERS132426
- SRA/ERS132427
- SRA/ERS132428
- SRA/ERS132429
- SRA/ERS132430
- SRA/ERS132431
- SRA/ERS132432
- SRA/ERS132433
- SRA/ERS132434
- SRA/ERS132435
- SRA/ERS132436
- SRA/ERS132437
- SRA/ERS132438
- SRA/ERS132439
- SRA/ERS132440
- SRA/ERS132441
- SRA/ERS132442
- SRA/ERS132443
- SRA/ERS132444
- SRA/ERS132445
- SRA/ERS132446
- SRA/ERS132447
- SRA/ERS132448
- SRA/ERS132449
- SRA/ERS132450
- SRA/ERS132451
- SRA/ERS132452
- SRA/ERS132453
- SRA/ERS132454
- SRA/ERS132456
- SRA/ERS132457
- SRA/ERS132458
- SRA/ERS132459
- SRA/ERS132460
- SRA/ERS132461
- SRA/ERS132462
- SRA/ERS132463
- SRA/ERS132464
- SRA/ERS132465
- SRA/ERS132466
- SRA/ERS132467
- SRA/ERS132468
- SRA/ERS132469
- SRA/ERS132471
- SRA/ERS132472
- SRA/ERS132473
- SRA/ERS132474
- SRA/ERS132475
- SRA/ERS132476
- SRA/ERS132477
- SRA/ERS132478
- SRA/ERS132479
- SRA/ERS132480
- SRA/ERS132481
- SRA/ERS132482
- SRA/ERS132483
- SRA/ERS132484
- SRA/ERS132485
- SRA/ERS132486
- SRA/ERS132487
- SRA/ERS132488
- SRA/ERS132489
- SRA/ERS132490
- SRA/ERS132491
- SRA/ERS132492
- SRA/ERS132493
- SRA/ERS132494
- SRA/ERS132495
- SRA/ERS132496
- SRA/ERS132497
- SRA/ERS132498
- SRA/ERS132499
- SRA/ERS132500
- SRA/ERS132502
- SRA/ERS132505
- SRA/ERS132506
- SRA/ERS132507
- SRA/ERS132508
- SRA/ERS132509
- SRA/ERS132510
- SRA/ERS132511
- SRA/ERS132512
- SRA/ERS132513
- SRA/ERS132514
- SRA/ERS132515
- SRA/ERS132516
- SRA/ERS132517
- SRA/ERS132518
- SRA/ERS132519
- SRA/ERS132520
- SRA/ERS132521
- SRA/ERS132522
- SRA/ERS132523
- SRA/ERS132524
- SRA/ERS132525
- SRA/ERS132526
- SRA/ERS132527
- SRA/ERS132528
- SRA/ERS132529
- SRA/ERS132530
- SRA/ERS132531
- SRA/ERS132532
- SRA/ERS132533
- SRA/ERS132534
- SRA/ERS132535
- SRA/ERS132536
- SRA/ERS132538
- SRA/ERS132539
- SRA/ERS132540
- SRA/ERS132541
- SRA/ERS132542
- SRA/ERS132543
- SRA/ERS132544
- SRA/ERS132545
- SRA/ERS132546
- SRA/ERS132547
- SRA/ERS132548
- SRA/ERS132549
- SRA/ERS132550
- SRA/ERS132551
- SRA/ERS156197
- SRA/ERS156198
- SRA/ERS156199
- SRA/ERS156200
- SRA/ERS156201
- SRA/ERS156202
- SRA/ERS156203
- SRA/ERS156204
- SRA/ERS156205
- SRA/ERS156206
- SRA/ERS156207
- SRA/ERS156208
- SRA/ERS156209
- SRA/ERS156210
- SRA/ERS156211
- SRA/ERS156212
- SRA/ERS156213
- SRA/ERS156214
- SRA/ERS156215
- SRA/ERS156216
- SRA/ERS156217
- SRA/ERS156218
- SRA/ERS156220
- SRA/ERS156221
- SRA/ERS156222
- SRA/ERS156223
- SRA/ERS156224
- SRA/ERS156225
- SRA/ERS156226
- SRA/ERS156227
- SRA/ERS156228
- SRA/ERS156229
- SRA/ERS156230
- SRA/ERS156231
- SRA/ERS156232
- SRA/ERS156233
- SRA/ERS156234
- SRA/ERS156235
- SRA/ERS156236
- SRA/ERS156237
- SRA/ERS156238
- SRA/ERS156239
- SRA/ERS156240
- SRA/ERS156241
- SRA/ERS156242
- SRA/ERS156243
- SRA/ERS156244
- SRA/ERS156245
- SRA/ERS156246
- SRA/ERS156247
- SRA/ERS156248
- SRA/ERS156249
- SRA/ERS156250
- SRA/ERS156251
- SRA/ERS156252
- SRA/ERS156253
- SRA/ERS156254
- SRA/ERS156255
- SRA/ERS156256
- SRA/ERS156257
- SRA/ERS156258
- SRA/ERS156259
- SRA/ERS156260
- SRA/ERS156261
- SRA/ERS156262
- SRA/ERS156263
- SRA/ERS156264
- SRA/ERS156265
- SRA/ERS156266
- SRA/ERS156267
- SRA/ERS156268
- SRA/ERS156269
- SRA/ERS156270
- SRA/ERS156271
- SRA/ERS156272
- SRA/ERS156273
- SRA/ERS156274
- SRA/ERS156275
- SRA/ERS156276
- SRA/ERS156277
- SRA/ERS156278
- SRA/ERS156279
- SRA/ERS156280
- SRA/ERS156281
- SRA/ERS156282
- SRA/ERS156283
- SRA/ERS156284
- SRA/ERS156285
- SRA/ERS156286
- SRA/ERS156287
- SRA/ERS156288
- SRA/ERS156289
- SRA/ERS156290
- SRA/ERS156292
- SRA/ERS156293
- SRA/ERS156294
- SRA/ERS156295
- SRA/ERS156296
- SRA/ERS156297
- SRA/ERS156298
- SRA/ERS156299
- SRA/ERS156300
- SRA/ERS156301
- SRA/ERS156302
- SRA/ERS156303
- SRA/ERS156304
- SRA/ERS156305
- SRA/ERS156306
- SRA/ERS156307
- SRA/ERS156308
- SRA/ERS156309
- SRA/ERS156310
- SRA/ERS156311
- SRA/ERS156312
- SRA/ERS156313
- SRA/ERS156314
- SRA/ERS156315
- SRA/ERS156316
- SRA/ERS156317
- SRA/ERS156318
- SRA/ERS156319
- SRA/ERS156320
- SRA/ERS156321
- SRA/ERS156322
- SRA/ERS156323
- SRA/ERS156324
- SRA/ERS156325
- SRA/ERS156326
- SRA/ERS156327
- SRA/ERS156328
- SRA/ERS156329
- SRA/ERS156330
- SRA/ERS156331
- SRA/ERS156332
- SRA/ERS156333
- SRA/ERS156334
- SRA/ERS156335
- SRA/ERS156336
- SRA/ERS156337
- SRA/ERS156338
- SRA/ERS156339
- SRA/ERS156340
- SRA/ERS156341
- SRA/ERS156342
- SRA/ERS156343
- SRA/ERS156344
- SRA/ERS156345
- SRA/ERS156346
- SRA/ERS156347
- SRA/ERS156348
- SRA/ERS156349
- SRA/ERS156350
- SRA/ERS156351
- SRA/ERS156352
- SRA/ERS156353
- SRA/ERS156354
- SRA/ERS156355
- SRA/ERS156356
- SRA/ERS156357
- SRA/ERS156358
- SRA/ERS156359
- SRA/ERS156360
- SRA/ERS156361
- SRA/ERS156362
- SRA/ERS156363
- SRA/ERS156364
- SRA/ERS156365
- SRA/ERS156366
- SRA/ERS156367
- SRA/ERS156368
- SRA/ERS156369
- SRA/ERS156370
- SRA/ERS156371
- SRA/ERS156372
- SRA/ERS156373
- SRA/ERS156374
- SRA/ERS156375
- SRA/ERS156376
- SRA/ERS156377
- SRA/ERS156378
- SRA/ERS156379
- SRA/ERS156380
- SRA/ERS156381
- SRA/ERS156382
- SRA/ERS156383
- SRA/ERS156384
- SRA/ERS156385
- SRA/ERS156386
- SRA/ERS156387
- SRA/ERS156388
Grants and funding
- 100087/WT_/Wellcome Trust/United Kingdom
- 089276/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- BB/G019177/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 089276/WT_/Wellcome Trust/United Kingdom
- 100891/WT_/Wellcome Trust/United Kingdom
- BB/G003203/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G018553/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G019274/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT/093724/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- MR/K010174/1/MRC_/Medical Research Council/United Kingdom
- BB/G020744/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

