The hexameric structure of the human mitochondrial replicative helicase Twinkle
- PMID: 25824949
- PMCID: PMC4417153
- DOI: 10.1093/nar/gkv189
The hexameric structure of the human mitochondrial replicative helicase Twinkle
Abstract
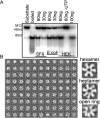
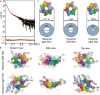
The mitochondrial replicative helicase Twinkle is involved in strand separation at the replication fork of mitochondrial DNA (mtDNA). Twinkle malfunction is associated with rare diseases that include late onset mitochondrial myopathies, neuromuscular disorders and fatal infantile mtDNA depletion syndrome. We examined its 3D structure by electron microscopy (EM) and small angle X-ray scattering (SAXS) and built the corresponding atomic models, which gave insight into the first molecular architecture of a full-length SF4 helicase that includes an N-terminal zinc-binding domain (ZBD), an intermediate RNA polymerase domain (RPD) and a RecA-like hexamerization C-terminal domain (CTD). The EM model of Twinkle reveals a hexameric two-layered ring comprising the ZBDs and RPDs in one layer and the CTDs in another. In the hexamer, contacts in trans with adjacent subunits occur between ZBDs and RPDs, and between RPDs and CTDs. The ZBDs show important structural heterogeneity. In solution, the scattering data are compatible with a mixture of extended hexa- and heptameric models in variable conformations. Overall, our structural data show a complex network of dynamic interactions that reconciles with the structural flexibility required for helicase activity.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. - PubMed
-
- Leipe D.D., Aravind L., Grishin N.V., Koonin E.V. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. - PubMed
-
- Shutt T.E., Gray M.W. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 2006;62:588–599. - PubMed
-
- Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

