Robust BRCA1-like classification of copy number profiles of samples repeated across different datasets and platforms
- PMID: 25825120
- PMCID: PMC5528812
- DOI: 10.1016/j.molonc.2015.03.002
Robust BRCA1-like classification of copy number profiles of samples repeated across different datasets and platforms
Abstract
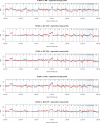
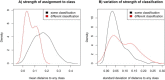
Breast cancers with BRCA1 germline mutation have a characteristic DNA copy number (CN) pattern. We developed a test that assigns CN profiles to be 'BRCA1-like' or 'non-BRCA1-like', which refers to resembling a BRCA1-mutated tumor or resembling a tumor without a BRCA1 mutation, respectively. Approximately one third of the BRCA1-like breast cancers have a BRCA1 mutation, one third has hypermethylation of the BRCA1 promoter and one third has an unknown reason for being BRCA1-like. This classification is indicative of patients' response to high dose alkylating and platinum containing chemotherapy regimens, which targets the inability of BRCA1 deficient cells to repair DNA double strand breaks. We investigated whether this classification can be reliably obtained with next generation sequencing and copy number platforms other than the bacterial artificial chromosome (BAC) array Comparative Genomic Hybridization (aCGH) on which it was originally developed. We investigated samples from 230 breast cancer patients for which a CN profile had been generated on two to five platforms, comprising low coverage CN sequencing, CN extraction from targeted sequencing panels (CopywriteR), Affymetrix SNP6.0, 135K/720K oligonucleotide aCGH, Affymetrix Oncoscan FFPE (MIP) technology, 3K BAC and 32K BAC aCGH. Pairwise comparison of genomic position-mapped profiles from the original aCGH platform and other platforms revealed concordance. For most cases, biological differences between samples exceeded the differences between platforms within one sample. We observed the same classification across different platforms in over 80% of the patients and kappa values of at least 0.36. Differential classification could be attributed to CN profiles that were not strongly associated to one class. In conclusion, we have shown that the genomic regions that define our BRCA1-like classifier are robustly measured by different CN profiling technologies, providing the possibility to retro- and prospectively investigate BRCA1-like classification across a wide range of CN platforms.
Keywords: BRCA1; Breast cancer; Classification; Copy number aberration profiles.
Copyright © 2015 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Aboyoun, P. , Pages, H. , Lawrence, M. , 2013. GenomicRanges: Representation and Manipulation of Genomic Intervals. R Package Version 1.12.5.
-
- Alvarez, S. , Diaz-Uriarte, R. , Osorio, A. , Barroso, A. , Melchor, L. , Paz, M.F. , Honrado, E. , Rodríguez, R. , Urioste, M. , Valle, L. , Díez, O. , Cigudosa, J.C. , Dopazo, J. , Esteller, M. , Benitez, J. , 2005. A predictor based on the somatic genomic changes of the BRCA1/BRCA2 breast cancer tumors identifies the non-BRCA1/BRCA2 tumors with BRCA1 promoter hypermethylation. Clin. Cancer Res. 11, 1146–1153. - PubMed
-
- Baumbusch, L.O. , Aarøe, J. , Johansen, F.E. , Hicks, J. , Sun, H. , Bruhn, L. , Gunderson, K. , Naume, B. , Kristensen, V.N. , Liestøl, K. , Børresen-Dale, A.-L. , Lingjaerde, O.C. , 2008. Comparison of the Agilent, ROMA/NimbleGen and Illumina platforms for classification of copy number alterations in human breast tumors. BMC Genomics 9, 379 - PMC - PubMed
-
- Bengtsson, H. , Irizarry, R. , Carvalho, B. , Speed, T.P. , 2008. Estimation and assessment of raw copy numbers at the single locus level. Bioinforma 24, 759–767. - PubMed
-
- Benjamini, Y. , Speed, T.P. , 2012. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res 40, e72 10.1093/nar/gks001 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

