Oxygen-Dependent Cell-to-Cell Variability in the Output of the Escherichia coli Tor Phosphorelay
- PMID: 25825431
- PMCID: PMC4438215
- DOI: 10.1128/JB.00074-15
Oxygen-Dependent Cell-to-Cell Variability in the Output of the Escherichia coli Tor Phosphorelay
Abstract
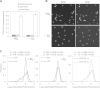
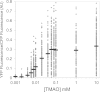
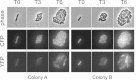
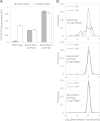
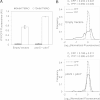
Escherichia coli senses and responds to trimethylamine-N-oxide (TMAO) in the environment through the TorT-TorS-TorR signal transduction system. The periplasmic protein TorT binds TMAO and stimulates the hybrid kinase TorS to phosphorylate the response regulator TorR through a phosphorelay. Phosphorylated TorR, in turn, activates transcription of the torCAD operon, which encodes the proteins required for anaerobic respiration via reduction of TMAO to trimethylamine. Interestingly, E. coli respires TMAO in both the presence and absence of oxygen, a behavior that is markedly different from the utilization of other alternative electron acceptors by this bacterium. Here we describe an unusual form of regulation by oxygen for this system. While the average level of torCAD transcription is the same for aerobic and anaerobic cultures containing TMAO, the behavior across the population of cells is strikingly different under the two growth conditions. Cellular levels of torCAD transcription in aerobic cultures are highly heterogeneous, in contrast to the relatively homogeneous distribution in anaerobic cultures. Thus, oxygen regulates the variance of the output but not the mean for the Tor system. We further show that this oxygen-dependent variability stems from the phosphorelay.
Importance: Trimethylamine-N-oxide (TMAO) is utilized by numerous bacteria as an electron acceptor for anaerobic respiration. In E. coli, expression of the proteins required for TMAO respiration is tightly regulated by a signal transduction system that is activated by TMAO. Curiously, although oxygen is the energetically preferred electron acceptor, TMAO is respired even in the presence of oxygen. Here we describe an interesting and unexpected form of regulation for this system in which oxygen produces highly variable expression of the TMAO utilization proteins across a population of cells without affecting the mean expression of these proteins. To our knowledge, this is the first reported example of a stimulus regulating the variance but not the mean output of a signaling system.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Comment in
-
Regulation by the modulation of gene expression variability.J Bacteriol. 2015 Jun 15;197(12):1974-5. doi: 10.1128/JB.00235-15. Epub 2015 Mar 30. J Bacteriol. 2015. PMID: 25825435 Free PMC article.
References
-
- Zeisel SH, daCosta KA, Youssef M, Hensey S. 1989. Conversion of dietary choline to trimethylamine and dimethylamine in rats: dose-response relationship. J Nutr 119:800–804. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

