Reduced Ssy1-Ptr3-Ssy5 (SPS) signaling extends replicative life span by enhancing NAD+ homeostasis in Saccharomyces cerevisiae
- PMID: 25825491
- PMCID: PMC4432292
- DOI: 10.1074/jbc.M115.644534
Reduced Ssy1-Ptr3-Ssy5 (SPS) signaling extends replicative life span by enhancing NAD+ homeostasis in Saccharomyces cerevisiae
Abstract
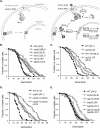
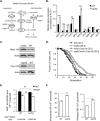
Attenuated nutrient signaling extends the life span in yeast and higher eukaryotes; however, the mechanisms are not completely understood. Here we identify the Ssy1-Ptr3-Ssy5 (SPS) amino acid sensing pathway as a novel longevity factor. A null mutation of SSY5 (ssy5Δ) increases replicative life span (RLS) by ∼50%. Our results demonstrate that several NAD(+) homeostasis factors play key roles in this life span extension. First, expression of the putative malate-pyruvate NADH shuttle increases in ssy5Δ cells, and deleting components of this shuttle, MAE1 and OAC1, largely abolishes RLS extension. Next, we show that Stp1, a transcription factor of the SPS pathway, directly binds to the promoter of MAE1 and OAC1 to regulate their expression. Additionally, deletion of SSY5 increases nicotinamide riboside (NR) levels and phosphate-responsive (PHO) signaling activity, suggesting that ssy5Δ increases NR salvaging. This increase contributes to NAD(+) homeostasis, partially ameliorating the NAD(+) deficiency and rescuing the short life span of the npt1Δ mutant. Moreover, we observed that vacuolar phosphatase, Pho8, is partially required for ssy5Δ-mediated NR increase and RLS extension. Together, our studies present evidence that supports SPS signaling is a novel NAD(+) homeostasis factor and ssy5Δ-mediated life span extension is likely due to concomitantly increased mitochondrial and vacuolar function. Our findings may contribute to understanding the molecular basis of NAD(+) metabolism, cellular life span, and diseases associated with NAD(+) deficiency and aging.
Keywords: NAD biosynthesis; cell metabolism; metabolic regulation; nicotinamide riboside salvage; yeast genetics; yeast metabolism.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Lin S. J., Defossez P. A., Guarente L. (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 - PubMed
-
- Fabrizio P., Pozza F., Pletcher S. D., Gendron C. M., Longo V. D. (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292, 288–290 - PubMed
-
- Kaeberlein M., Powers R. W., 3rd, Steffen K. K., Westman E. A., Hu D., Dang N., Kerr E. O., Kirkland K. T., Fields S., Kennedy B. K. (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

