A novel role of farnesylation in targeting a mitotic checkpoint protein, human Spindly, to kinetochores
- PMID: 25825516
- PMCID: PMC4384735
- DOI: 10.1083/jcb.201412085
A novel role of farnesylation in targeting a mitotic checkpoint protein, human Spindly, to kinetochores
Abstract
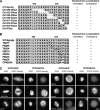
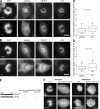
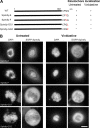
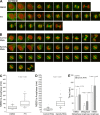
Kinetochore (KT) localization of mitotic checkpoint proteins is essential for their function during mitosis. hSpindly KT localization is dependent on the RZZ complex and hSpindly recruits the dynein-dynactin complex to KTs during mitosis, but the mechanism of hSpindly KT recruitment is unknown. Through domain-mapping studies we characterized the KT localization domain of hSpindly and discovered it undergoes farnesylation at the C-terminal cysteine residue. The N-terminal 293 residues of hSpindly are dispensable for its KT localization. Inhibition of farnesylation using a farnesyl transferase inhibitor (FTI) abrogated hSpindly KT localization without affecting RZZ complex, CENP-E, and CENP-F KT localization. We showed that hSpindly is farnesylated in vivo and farnesylation is essential for its interaction with the RZZ complex and hence KT localization. FTI treatment and hSpindly knockdown displayed the same mitotic phenotypes, indicating that hSpindly is a key FTI target in mitosis. Our data show a novel role of lipidation in targeting a checkpoint protein to KTs through protein-protein interaction.
© 2015 Moudgil et al.
Figures




References
-
- Ashar H.R., James L., Gray K., Carr D., Black S., Armstrong L., Bishop W.R., and Kirschmeier P.. 2000. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J. Biol. Chem. 275:30451–30457 10.1074/jbc.M003469200 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

