Autoinhibition of MDMX by intramolecular p53 mimicry
- PMID: 25825738
- PMCID: PMC4403185
- DOI: 10.1073/pnas.1420833112
Autoinhibition of MDMX by intramolecular p53 mimicry
Abstract
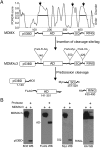
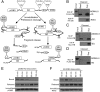
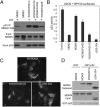
The p53 inhibitor MDMX is controlled by multiple stress signaling pathways. Using a proteolytic fragment release (PFR) assay, we detected an intramolecular interaction in MDMX that mechanistically mimics the interaction with p53, resulting in autoinhibition of MDMX. This mimicry is mediated by a hydrophobic peptide located in a long disordered central segment of MDMX that has sequence similarity to the p53 transactivation domain. NMR spectroscopy was used to show this hydrophobic peptide interacts with the N-terminal domain of MDMX in a structurally analogous manner to p53. Mutation of two critical tryptophan residues in the hydrophobic peptide disrupted the intramolecular interaction and increased p53 binding, providing further evidence for mechanistic mimicry. The PFR assay also revealed a second intramolecular interaction between the RING domain and central region that regulates MDMX nuclear import. These results establish the importance of intramolecular interactions in MDMX regulation, and validate a new assay for the study of intramolecular interactions in multidomain proteins with intrinsically disordered regions.
Keywords: MDMX; acidic domain; intramolecular; p53; protease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

