Integrated in silico Analyses of Regulatory and Metabolic Networks of Synechococcus sp. PCC 7002 Reveal Relationships between Gene Centrality and Essentiality
- PMID: 25826650
- PMCID: PMC4500133
- DOI: 10.3390/life5021127
Integrated in silico Analyses of Regulatory and Metabolic Networks of Synechococcus sp. PCC 7002 Reveal Relationships between Gene Centrality and Essentiality
Abstract
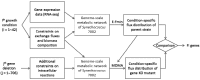
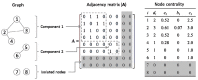
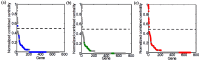
Cyanobacteria dynamically relay environmental inputs to intracellular adaptations through a coordinated adjustment of photosynthetic efficiency and carbon processing rates. The output of such adaptations is reflected through changes in transcriptional patterns and metabolic flux distributions that ultimately define growth strategy. To address interrelationships between metabolism and regulation, we performed integrative analyses of metabolic and gene co-expression networks in a model cyanobacterium, Synechococcus sp. PCC 7002. Centrality analyses using the gene co-expression network identified a set of key genes, which were defined here as "topologically important." Parallel in silico gene knock-out simulations, using the genome-scale metabolic network, classified what we termed as "functionally important" genes, deletion of which affected growth or metabolism. A strong positive correlation was observed between topologically and functionally important genes. Functionally important genes exhibited variable levels of topological centrality; however, the majority of topologically central genes were found to be functionally essential for growth. Subsequent functional enrichment analysis revealed that both functionally and topologically important genes in Synechococcus sp. PCC 7002 are predominantly associated with translation and energy metabolism, two cellular processes critical for growth. This research demonstrates how synergistic network-level analyses can be used for reconciliation of metabolic and gene expression data to uncover fundamental biological principles.
Figures




References
-
- Bernstein H.C., Konopka A., Melnicki M.R., Hill E.A., Kucek L.A., Zhang S.Y., Shen G.Z., Bryant D.A., Beliaev A.S. Effect of mono- and dichromatic light quality on growth rates and photosynthetic performance of synechococcus sp pcc 7002. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00488. - DOI - PMC - PubMed
-
- Grossman A.R., Schaefer M.R., Chiang G.G., Collier J.L. The Molecular Biology of Cyanobacteria. Springer; Berlin/Heidelberg, Germany: 2004. The responses of cyanobacteria to environmental conditions: Light and nutrients; pp. 641–675.
LinkOut - more resources
Full Text Sources
Other Literature Sources

