dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants
- PMID: 25827226
- PMCID: PMC4434706
- DOI: 10.1073/pnas.1422948112
dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants
Abstract
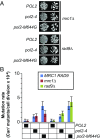
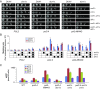
Mutator phenotypes create genetic diversity that fuels tumor evolution. DNA polymerase (Pol) ε mediates leading strand DNA replication. Proofreading defects in this enzyme drive a number of human malignancies. Here, using budding yeast, we show that mutator variants of Pol ε depend on damage uninducible (Dun)1, an S-phase checkpoint kinase that maintains dNTP levels during a normal cell cycle and up-regulates dNTP synthesis upon checkpoint activation. Deletion of DUN1 (dun1Δ) suppresses the mutator phenotype of pol2-4 (encoding Pol ε proofreading deficiency) and is synthetically lethal with pol2-M644G (encoding altered Pol ε base selectivity). Although pol2-4 cells cycle normally, pol2-M644G cells progress slowly through S-phase. The pol2-M644G cells tolerate deletions of mediator of the replication checkpoint (MRC) 1 (mrc1Δ) and radiation sensitive (Rad) 9 (rad9Δ), which encode mediators of checkpoint responses to replication stress and DNA damage, respectively. The pol2-M644G mutator phenotype is partially suppressed by mrc1Δ but not rad9Δ; neither deletion suppresses the pol2-4 mutator phenotype. Thus, checkpoint activation augments the Dun1 effect on replication fidelity but is not required for it. Deletions of genes encoding key Dun1 targets that negatively regulate dNTP synthesis, suppress the dun1Δ pol2-M644G synthetic lethality and restore the mutator phenotype of pol2-4 in dun1Δ cells. DUN1 pol2-M644G cells have constitutively high dNTP levels, consistent with checkpoint activation. In contrast, pol2-4 and POL2 cells have similar dNTP levels, which decline in the absence of Dun1 and rise in the absence of the negative regulators of dNTP synthesis. Thus, dNTP pool levels correlate with Pol ε mutator severity, suggesting that treatments targeting dNTP pools could modulate mutator phenotypes for therapy.
Keywords: DNA replication and repair; cancer; lethal mutagenesis; polymerase fidelity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Pools and Pols: Mechanism of a mutator phenotype.Proc Natl Acad Sci U S A. 2015 May 12;112(19):5864-5. doi: 10.1073/pnas.1505169112. Epub 2015 Apr 30. Proc Natl Acad Sci U S A. 2015. PMID: 25931524 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

