Rational design of allosteric-inhibition sites in classical protein tyrosine phosphatases
- PMID: 25828055
- PMCID: PMC4451255
- DOI: 10.1016/j.bmc.2015.03.027
Rational design of allosteric-inhibition sites in classical protein tyrosine phosphatases
Abstract
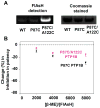
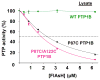
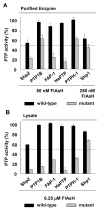
Protein tyrosine phosphatases (PTPs), which catalyze the dephosphorylation of phosphotyrosine in protein substrates, are critical regulators of metazoan cell signaling and have emerged as potential drug targets for a range of human diseases. Strategies for chemically targeting the function of individual PTPs selectively could serve to elucidate the signaling roles of these enzymes and would potentially expedite validation of the therapeutic promise of PTP inhibitors. Here we report a novel strategy for the design of non-natural allosteric-inhibition sites in PTPs; these sites, which can be introduced into target PTPs through protein engineering, serve to sensitize target PTPs to potent and selective inhibition by a biarsenical small molecule. Building on the recent discovery of a naturally occurring cryptic allosteric site in wild-type Src-homology-2 domain containing PTP (Shp2) that can be targeted by biarsenical compounds, we hypothesized that Shp2's unusual sensitivity to biarsenicals could be strengthened through rational design and that the Shp2-specific site could serve as a blueprint for the introduction of non-natural inhibitor sensitivity in other PTPs. Indeed, we show here that the strategic introduction of a cysteine residue at a position removed from the Shp2 active site can serve to increase the potency and selectivity of the interaction between Shp2's allosteric site and the biarsenical inhibitor. Moreover, we find that 'Shp2-like' allosteric sites can be installed de novo in PTP enzymes that do not possess naturally occurring sensitivity to biarsenical compounds. Using primary-sequence alignments to guide our enzyme engineering, we have successfully introduced allosteric-inhibition sites in four classical PTPs-PTP1B, PTPH-1, FAP-1, and HePTP-from four different PTP subfamilies, suggesting that our sensitization approach can likely be applied widely across the classical PTP family to generate biarsenical-responsive PTPs.
Keywords: Allostery; Biarsenicals; FlAsH; Inhibitor sensitization; Protein engineering; Protein tyrosine phosphatases.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
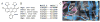

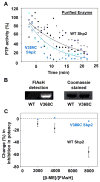

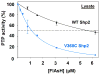
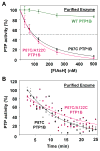
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

