Role of nucleotide binding and GTPase domain dimerization in dynamin-like myxovirus resistance protein A for GTPase activation and antiviral activity
- PMID: 25829498
- PMCID: PMC4432294
- DOI: 10.1074/jbc.M115.650325
Role of nucleotide binding and GTPase domain dimerization in dynamin-like myxovirus resistance protein A for GTPase activation and antiviral activity
Abstract
Myxovirus resistance (Mx) GTPases are induced by interferon and inhibit multiple viruses, including influenza and human immunodeficiency viruses. They have the characteristic domain architecture of dynamin-related proteins with an N-terminal GTPase (G) domain, a bundle signaling element, and a C-terminal stalk responsible for self-assembly and effector functions. Human MxA (also called MX1) is expressed in the cytoplasm and is partly associated with membranes of the smooth endoplasmic reticulum. It shows a protein concentration-dependent increase in GTPase activity, indicating regulation of GTP hydrolysis via G domain dimerization. Here, we characterized a panel of G domain mutants in MxA to clarify the role of GTP binding and the importance of the G domain interface for the catalytic and antiviral function of MxA. Residues in the catalytic center of MxA and the nucleotide itself were essential for G domain dimerization and catalytic activation. In pulldown experiments, MxA recognized Thogoto virus nucleocapsid proteins independently of nucleotide binding. However, both nucleotide binding and hydrolysis were required for the antiviral activity against Thogoto, influenza, and La Crosse viruses. We further demonstrate that GTP binding facilitates formation of stable MxA assemblies associated with endoplasmic reticulum membranes, whereas nucleotide hydrolysis promotes dynamic redistribution of MxA from cellular membranes to viral targets. Our study highlights the role of nucleotide binding and hydrolysis for the intracellular dynamics of MxA during its antiviral action.
Keywords: G protein; Mx proteins; antiviral response; catalytic mechanism; dynamin-like protein; innate immunity; interferon; membrane; viral replication.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

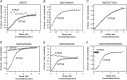

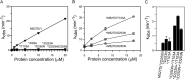
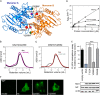
 ), M527D/D250N (XTP) (Δ), M527D/D253N (GTP) (□), and M527D/D250N/D253N (XTP) (○). B, GTPase activity of M527D can be stimulated by monomeric G domain mutants of MxA. 2.5 μ
), M527D/D250N (XTP) (Δ), M527D/D253N (GTP) (□), and M527D/D250N/D253N (XTP) (○). B, GTPase activity of M527D can be stimulated by monomeric G domain mutants of MxA. 2.5 μ
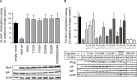
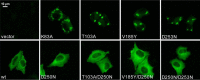
References
-
- Haller O., Staeheli P., Schwemmle M., Kochs G. (2015) Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 23, 154–163 - PubMed
-
- Haller O., Kochs G. (2011) Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interferon Cytokine Res. 31, 79–87 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

