Somatic cancer mutations in the MLL3-SET domain alter the catalytic properties of the enzyme
- PMID: 25829971
- PMCID: PMC4379744
- DOI: 10.1186/s13148-015-0075-3
Somatic cancer mutations in the MLL3-SET domain alter the catalytic properties of the enzyme
Abstract
Background: Somatic mutations in epigenetic enzymes are frequently found in cancer tissues. The MLL3 H3K4-specific protein lysine monomethyltransferase is an important epigenetic enzyme, and it is among the most recurrently mutated enzymes in cancers. MLL3 mainly introduces H3K4me1 at enhancers.
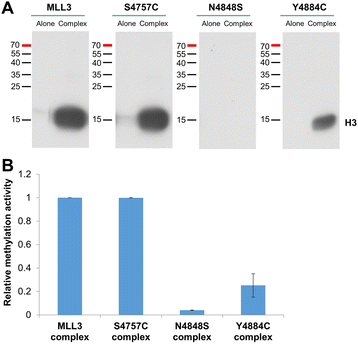
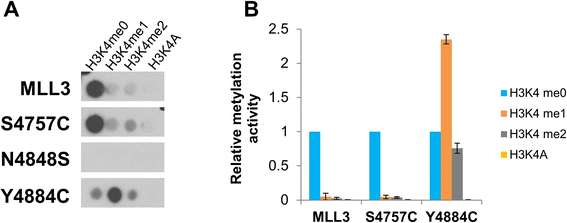
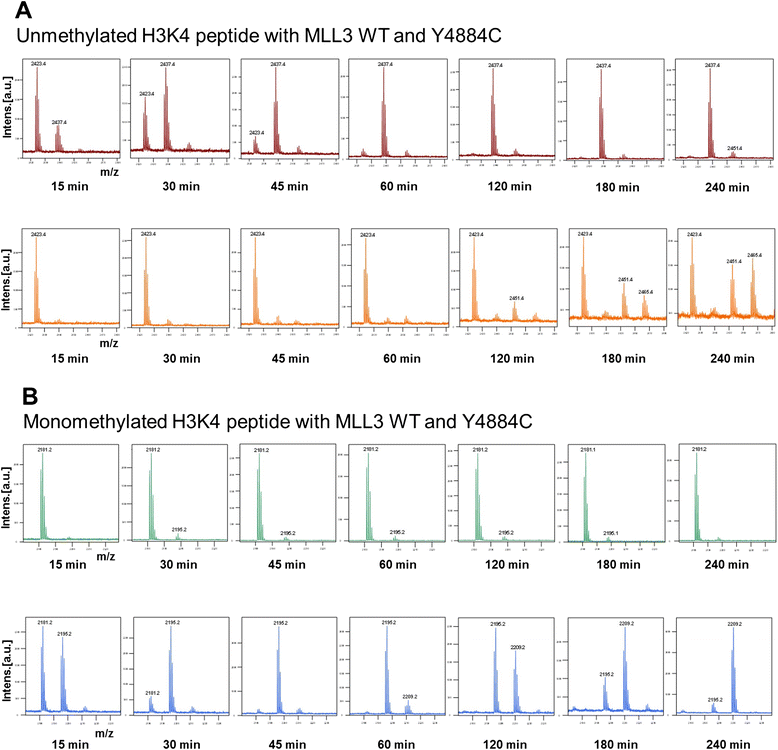
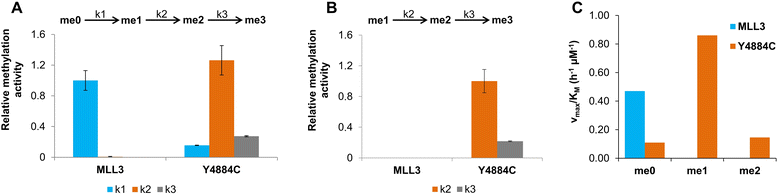
Results: We investigated the enzymatic properties of MLL3 variants that carry somatic cancer mutations. Asn4848 is located at the cofactor binding sites, and the N4848S exchange renders the enzyme inactive. Tyr4884 is part of an aromatic pocket at the active center of the enzyme, and Y4884C converts MLL3 from a monomethyltransferase with substrate preference for H3K4me0 to a trimethyltransferase with H3K4me1 as preferred substrate. Expression of Y4884C leads to aberrant H3K4me3 formation in cells.
Conclusions: Our data show that different somatic cancer mutations of MLL3 affect the enzyme activity in distinct and opposing manner highlighting the importance of experimentally studying the effects of somatic cancer mutations in key regulatory enzymes in order to develop and apply targeted tumor therapy.
Figures


Similar articles
-
Somatic mutations of the mixed-lineage leukemia 3 (MLL3) gene in primary breast cancers.Pathol Oncol Res. 2011 Jun;17(2):429-33. doi: 10.1007/s12253-010-9316-0. Epub 2010 Nov 30. Pathol Oncol Res. 2011. PMID: 21116761
-
MLL3/MLL4 enzymatic activity shapes DNA replication timing.bioRxiv [Preprint]. 2023 Dec 8:2023.12.07.569680. doi: 10.1101/2023.12.07.569680. bioRxiv. 2023. PMID: 38106216 Free PMC article. Preprint.
-
Therapeutic targeting of metabolic vulnerabilities in cancers with MLL3/4-COMPASS epigenetic regulator mutations.J Clin Invest. 2023 Jul 3;133(13):e169993. doi: 10.1172/JCI169993. J Clin Invest. 2023. PMID: 37252797 Free PMC article.
-
Pax6 associates with H3K4-specific histone methyltransferases Mll1, Mll2, and Set1a and regulates H3K4 methylation at promoters and enhancers.Epigenetics Chromatin. 2016 Sep 9;9(1):37. doi: 10.1186/s13072-016-0087-z. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27617035 Free PMC article.
-
A historical overview of protein kinases and their targeted small molecule inhibitors.Pharmacol Res. 2015 Oct;100:1-23. doi: 10.1016/j.phrs.2015.07.010. Epub 2015 Jul 21. Pharmacol Res. 2015. PMID: 26207888 Review.
Cited by
-
The importance of detailed epigenomic profiling of different cell types within organs.Epigenomics. 2016 Jun;8(6):817-29. doi: 10.2217/epi-2016-0005. Epub 2016 Jun 15. Epigenomics. 2016. PMID: 27305639 Free PMC article. Review.
-
H2B ubiquitylation enhances H3K4 methylation activities of human KMT2 family complexes.Nucleic Acids Res. 2020 Jun 4;48(10):5442-5456. doi: 10.1093/nar/gkaa317. Nucleic Acids Res. 2020. PMID: 32365172 Free PMC article.
-
NGS Analysis Confirms Common TP53 and RB1 Mutations, and Suggests MYC Amplification in Ocular Adnexal Sebaceous Carcinomas.Int J Mol Sci. 2021 Aug 6;22(16):8454. doi: 10.3390/ijms22168454. Int J Mol Sci. 2021. PMID: 34445161 Free PMC article.
-
Gain-of-Function Genetic Alterations of G9a Drive Oncogenesis.Cancer Discov. 2020 Jul;10(7):980-997. doi: 10.1158/2159-8290.CD-19-0532. Epub 2020 Apr 8. Cancer Discov. 2020. PMID: 32269030 Free PMC article.
-
COMPASS Ascending: Emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer.Cancer Lett. 2019 Aug 28;458:56-65. doi: 10.1016/j.canlet.2019.05.024. Epub 2019 May 22. Cancer Lett. 2019. PMID: 31128216 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

